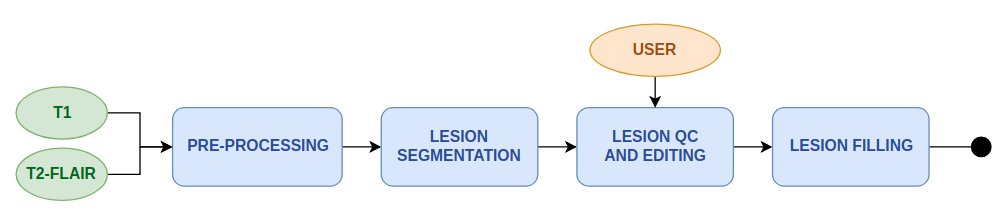
Lesion Segmentation Workflow
This workflow performs automatic segmentation of T2-FLAIR hyperintensity lesions and uses this information to "fill" the white matter lesions in a corresponding T1 image with surrounding healthy WM values. This enables a fast an automatic process to obtain lesions mask in T1 space, offers the possibility for semi-automatic or manual QC and editing of this lesion mask, and produces a lesion-filled T1 that can be used for improved quantitative calculations in morphometry and connectivity workflows, avoiding incorrect tissue segmentation assignment of the T1 lesions due to lower intensity values.
The workflow includes the following steps:
- Image pre-processing including Bias field correction of T1 image, AC-PC alignment, and rigid registration of T2-FLAIR to T1 (see Anatomy pre-processing)
- Automatic lesion segmentation from T2-FLAIR hyperintensities (see Lesion Segmentation Tool - LPA)
- Semi-automatic lesion mask QC and editing (see Manual & semi-automatic lesion-mask editing)
- Automatic lesion-filling of lesions in the T1 image with values of surrounding healthy tissue (see Lesion Filling)
Workflow scheme
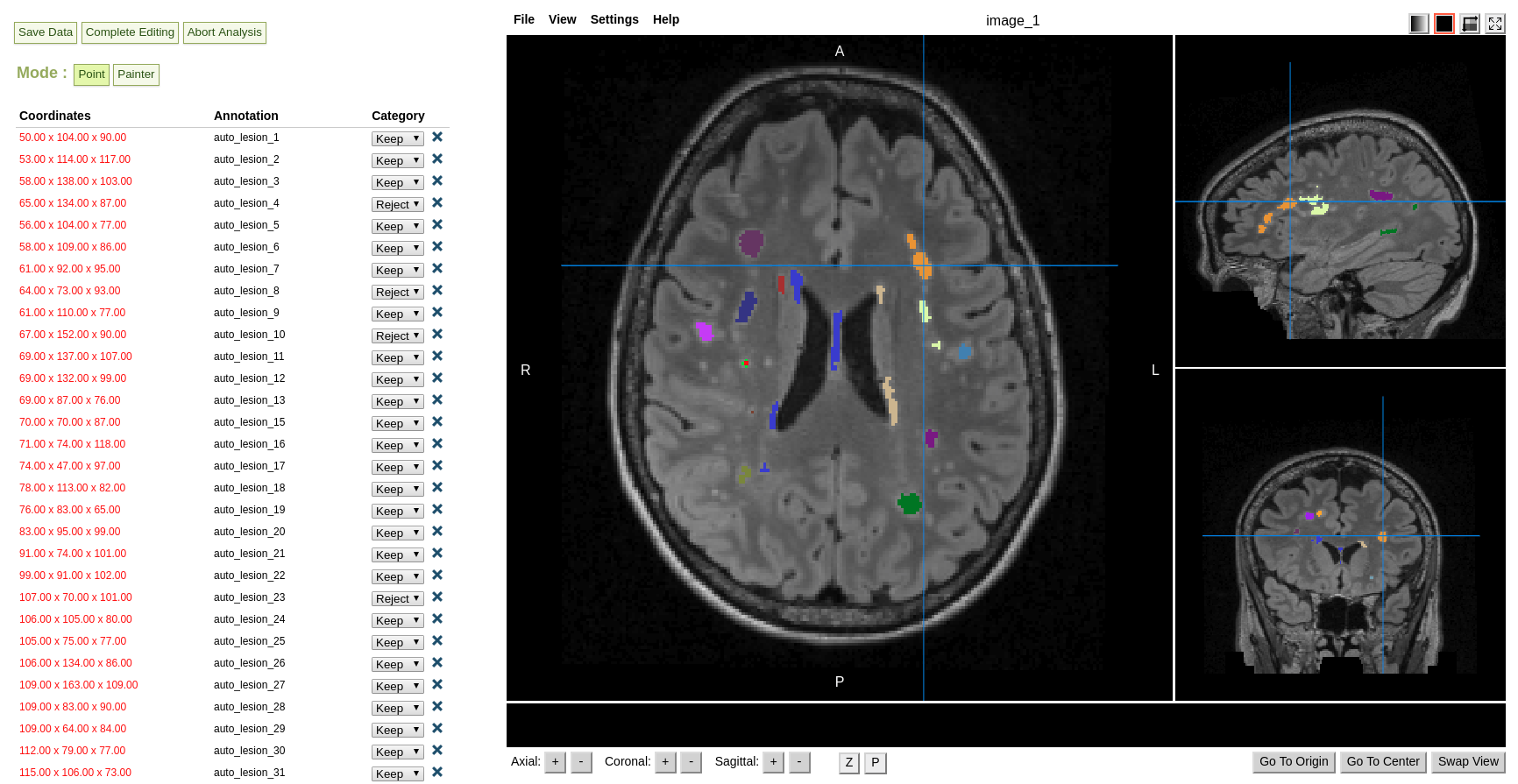
Lesion Segmentation Tool output examples
T2 FLAIR input |
Lesion probability mask |
Lesion segmentation |
Semi automatic lesion editing
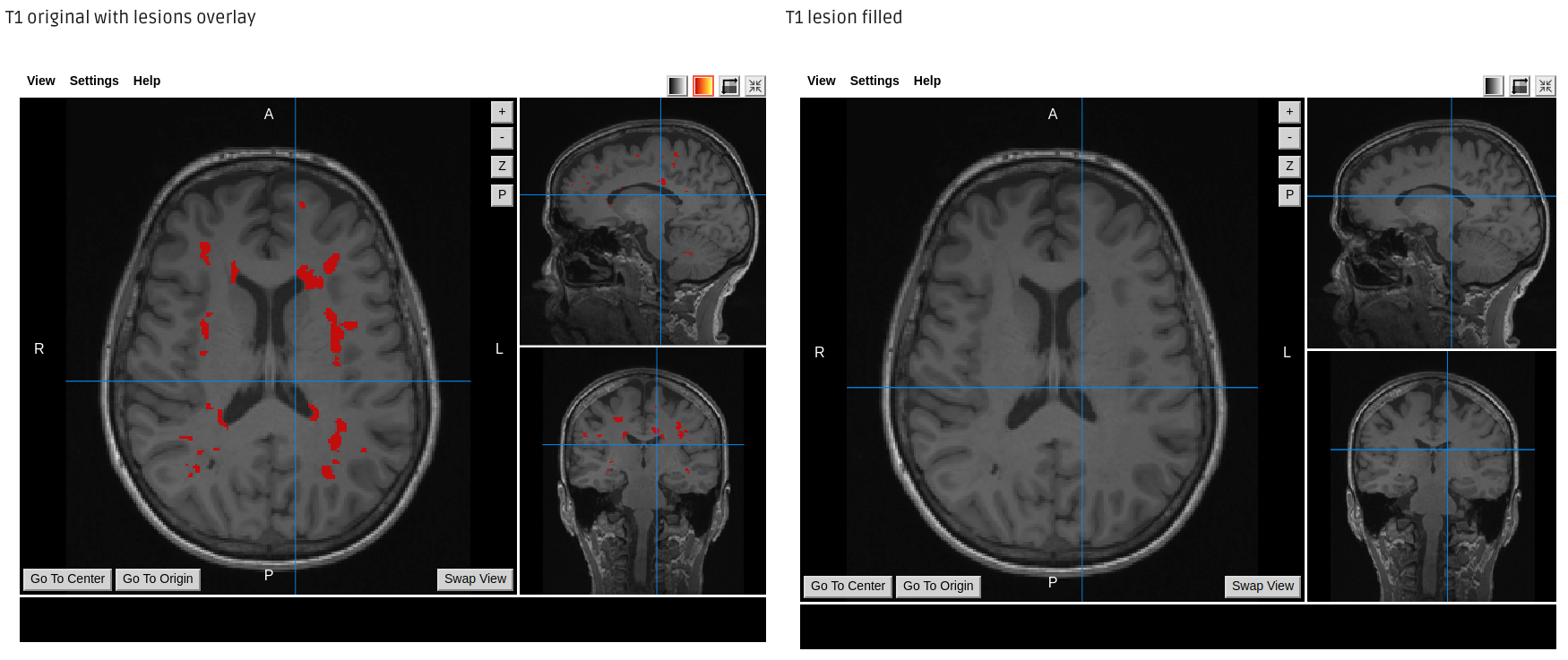
Lesion Filling output example
Required inputs:
- T1: anatomical 3D image to be lesion-filled.
Isotropic resolution recommended
Must be labelled as 'T1' modality - T2-FLAIR: anatomical 3D image with hyper-intense lesions
Isotropic resolution recommended
Must be labelled as 'T2' modality
Minimum input requirements:
- For optimal result reliability, isotropic resolution is recommended
- Recommended resolution: 1 mm isotropic.
- Minimum reliable resolution: 2mm isotropic.
References:
-
Refer to the following tool pages for relevant references:
Create free account now!
