fMRI structural connectome
Whole brain functional connectivity of a subject based on anatomical (T1 image) and resting-state functional data (rs-fMRI 4D time-series). This tool follows the scheme described by Hagmann et al. 2008.
The tool measures functional activity correlations between the different cortical and sub-cortical gray matter regions. the workflow includes full anatomical and functional image preprocessing (such as motion, bias-field and slice-timing correction, image co-registration, intenstiy normalization and band-pass filtering), full morphometric T1 analysis (including tissue segmentation and atlas-based parcellation) and connectomics analysis including graph-based network measures. This type of analysis is very useful in neurodegenrative diseases, pyschiatric pathology and neuroplasticity research, detecting differences in brain connectivity between patients and controls and showing areas or subnetworks affetced by a particular disease.
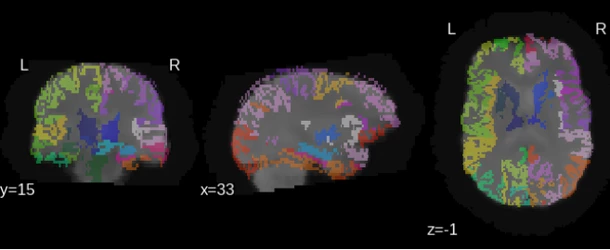
The regions of interest (ROIs) are computed by performing a morphometric analysis in the input T1 image, which may be done either by Custom morphology or FreeSurfer tools. This section of the analysis includes image pre-processing, tissue segmentation, atlas-based parcellation and labeling, and volumetric analysis. For more information visit the Custom morphology/ FreeSurfer help pages.
The functional data is pre-processed (movement correction, slice timing correction, brain extraction, smoothing, intensity normalization, and band-pass filtering) and the average signal intensity of every ROI is computed for every time-point (after region dilation as described by [Hagmann et al. 2008]). The resulting time-series are de-trended and the global brain signal is regressed out before computing cross-correlation maps.
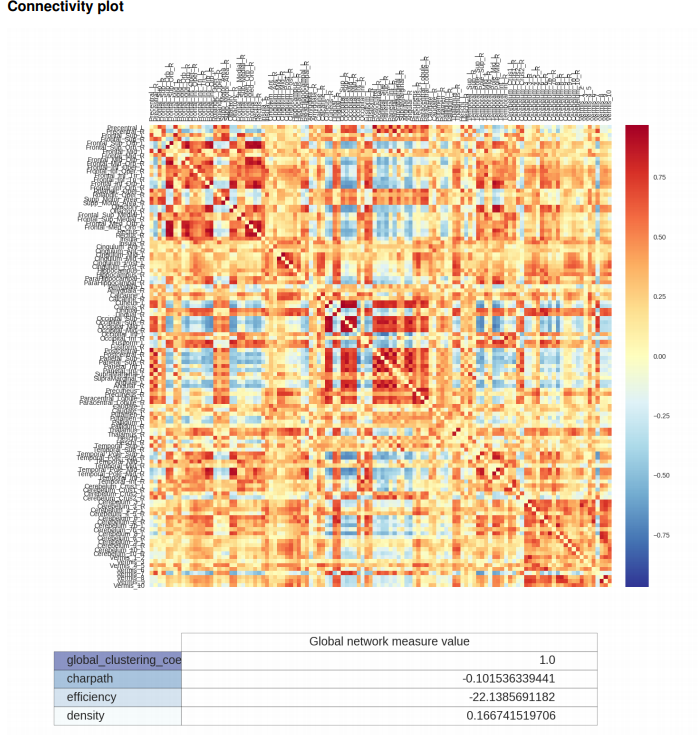
The final correlation information is stored in a functional connectivity matrix representing the strength of functional connection across pairs of regions.
rs-fMRI Connectome output examples
Connectivity matrix
Required inputs:
- T1: anatomical 3D image
Isotropic resolution recommended
Must be labeled as 'T1' modality - fMRI: resting-state functional 4D time series
Isotropic resolution recommended
Must be labeled as 'fMRI' modality"
Minimum input requirements:
- For optimal result reliability, isotropic resolution is recommended
- T1 recommended resolution: 1 mm isotropic.
- T1 minimum reliable resolution: 2mm isotropic.
- fMRI recommended resolution: 2-3 mm isotropic.
- fMRI minimum reliable resolution: 4 mm isotropic.
Settings:
- Pre-processing
- Preferred DICOM to NIfTI conversion tool (drop-down selection)
The selected tool will be tried first to convert DICOM to NIfTI. If the conversion fails, the other options will be tried sequentially until a successful conversion.- Mrtrix (Default)
- bru2nii
- DCM2nii
- mcverter
- diffunpack
- Preferred DICOM to NIfTI conversion tool (drop-down selection)
- Alignment
- Do ACPC alignment (checkbox)If checked do ACPC alignment (Default checked)
- Select tool for ACPC alignment (drop-down selection)
- ANTs (default)
- FSL
- Morphology tool
- Morphology tool (drop-down selection)
- Custom morphology (Default)
(selectable skull-stripping ; ANTs tissue segmentation; selectable parcellation) - FreeSurfer
- Custom morphology (Default)
- Atropos (ANTs) segmentation parameters (string)
- Default: -m 2 -n 20 -w 0.25
- Morphology tool (drop-down selection)
- Custom morphology options
Only if using Custom morphology option, skip for FreeSurfer option
- Input is already skull-stripped (checkbox)
If checked skips the brain-extraction step (Default unchecked). - T1 Skull-stripping tool (drop-down selection)
- Additional skull-stripping parameters (string)
String with additional command parameters for the skull stripping tool. Optional (Default empty).
- Brain Atlas for parcellation (drop-down selection)
Template and labeled atlas used to parcellate grey matter regions.- DKT40 (Mindboggle-101) (Default)
- AAL
- LPBA40
- Do thickness computation (checkbox)
- If checked computes cortical thickness measures. Significantly augments computation time (Default not checked).
- Input is already skull-stripped (checkbox)
- Functional time series processing
- Skip rs-fMRI preprocessing (checkbox)
If checked does not pre-process functional data (Default not checked)
- Global nuisance regression (checkbox)
If checked performs regression of the global signal to the functional timeseries data (Default checked)
- Skip rs-fMRI preprocessing (checkbox)
Outputs:
-
Report:
- report.pdf: report file with results summary.
-
Images:
- T1_original.nii.gz: the input T1 image used in the tool.
- T1_acpc.nii.gz: preprocessed T1 image acpc-aligned.
- T1mask.nii.gz: binary brain-extraction mask for the T1 image.
- T1strip.nii.gz: preprocessed and skull-stripped T1 image.
- T1_act.nii.gz: image containing the different segmented tissues as separate volumes.
- tissueSegmentation.nii.gz: 5-tissue (csf, gm, wm, brainstem, cerebellum) tissue segmentation image.
- atlas_registered.nii.gz: template atlas labels registered into subject space.
- full_labeled.nii.gz: subject parcellated labels image, using all labels provided by the selected atlas.
- labeled.nii.gz: subject label image with a selection of labels from of 85 relevant gray matter regions.
- fMRI.nii.gz: input functional image used in the tool.
- filtered_func_data.nii.gz: preprocessed functional data.
- filtered_func_data_res.nii.gz: preprocessed functional data after nuisance regression.
- fmri_mean.nii.gz: average of all functional volumes.
- T1_to_func0GenericAffine.mat: affine registration matrix from T1 to fMRI.
- T1_to_funcWarped.nii.gz: T1 non-linearly warped to the fMRI space.
- T1mask_to_func.nii.gz: mask warped to functional space
- labeled_to_func.nii.gz: anatomical regions warped to functional space.
- tissueSegmentation_to_func.nii.gz: tissue segmentation warped to functional space.
-
Datasheets:
- resting_cmp_matrix.csv: functional connectome matrix.
- tissueSegmentation.csv: comma-separated-value (CSV) file with tissue types volume information How to interpret volumetry .csv outputs .
- volumetric.csv: CSV file with gray matter parcellated regions volume and cortical thickness information (includes tissue types) How to interpret volumetry .csv outputs .
-
Folders:
- FreeSurfer: Folder with full outputs of the FreeSurfer's recon-all script (if FreeSurfer option chosen).
Typical execution time:
- 1 hour (with default parameters).
- 6 hours (using FreeSurfer option).
References:
- Overall scheme: Hagmann et al. 2008
- Nuisance regression: Saad et al. 2012
- FSL fMRI preprocessing tools: Smith et al. 2004
- Refer to Custom morphology/FreeSurfer for relevant references when using the respective option.
Create free account now!
