Custom morphology
Full morphology processing of a T1 anatomical image using an array of tools where the user can customize the preferred tool for each step of the pipeline.
This tool can use age-matched templates for tissue and region parcellation priors – using the information from the Age-at-scan field – for subjects between 3 months and 17 years-old, making it especially suited for children and young adult data.
The morphology pipeline Includes the following steps:
- Skull-stripping
- Bias-field correction
- AC-PC alignment to template.
- Tissue segmentation into CSF, gray matter and white matter.
- Atlas-based parcellation and labeling using LPBA40 56-region parcellation scheme Shattuck et al. 2008 .
- Volumetry (for each tissue type and parcellated regions).
- Cortical thickness computation for cortical regions.
Custom Morphology output examples
Grey Matter Labels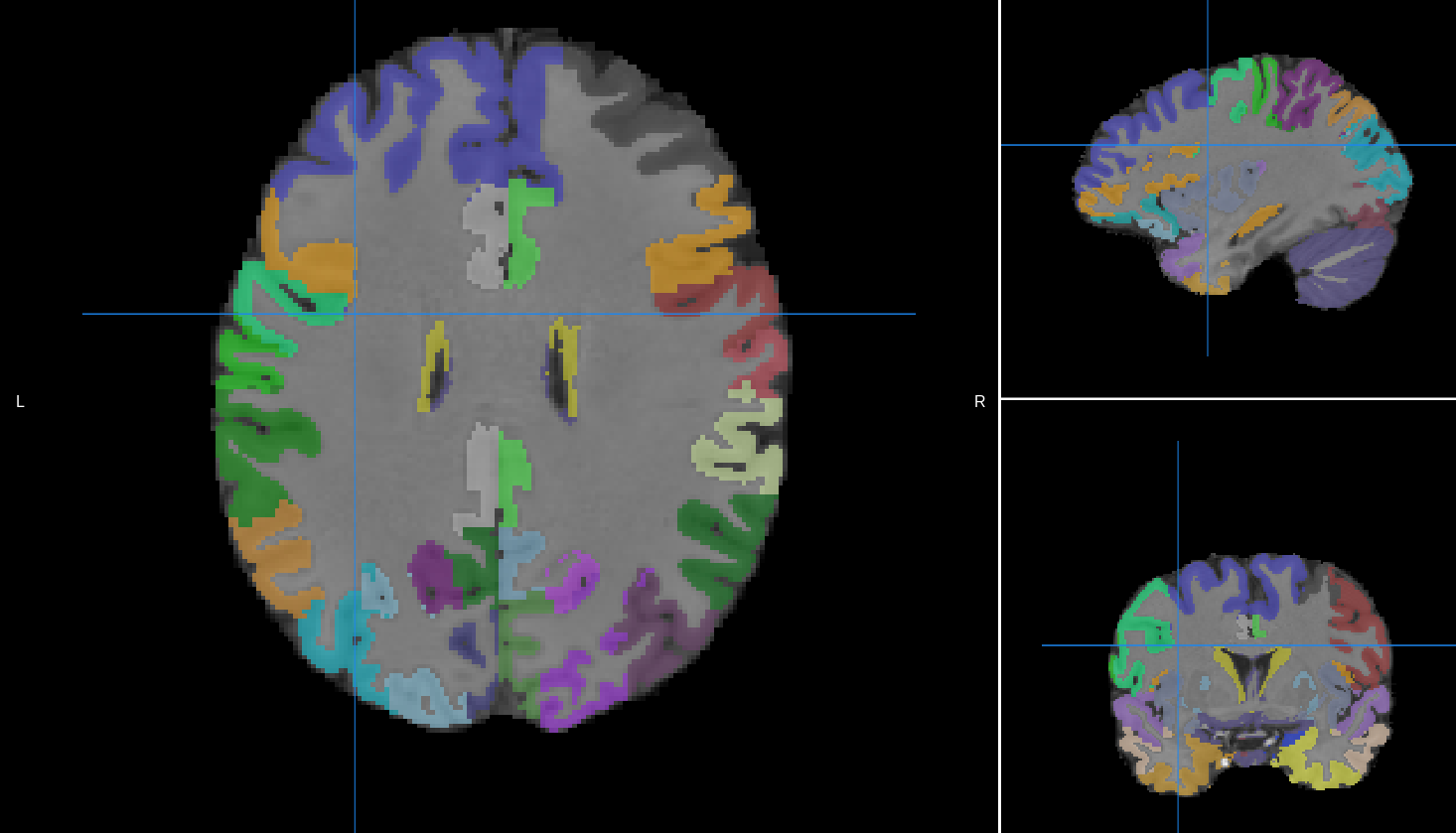 |
Cortical Thickness 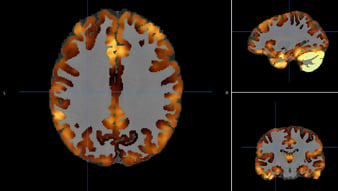 |
Analysis Report
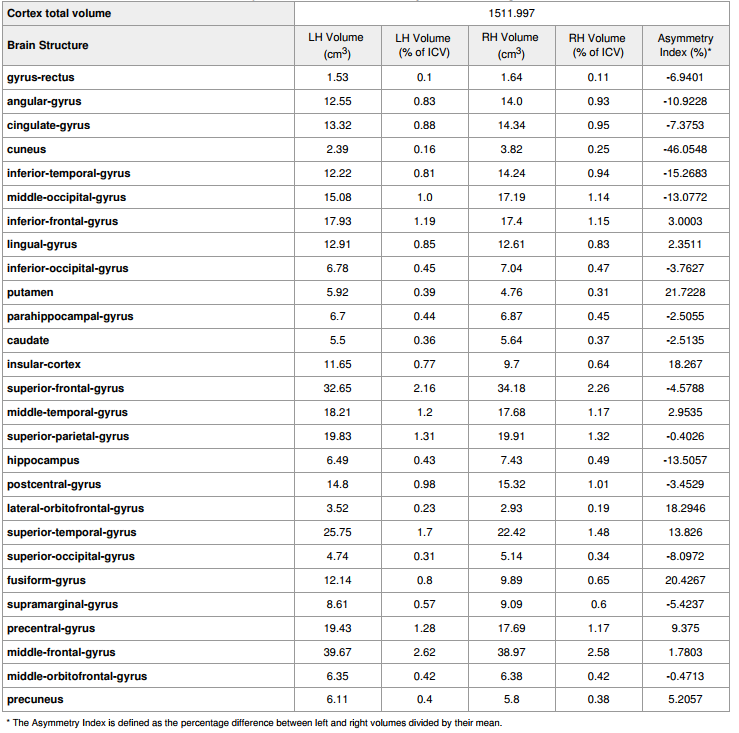 |
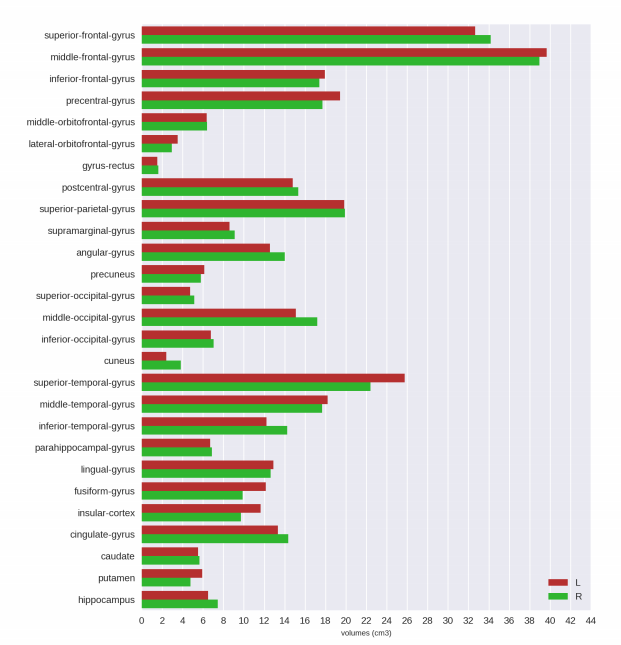 |
Required inputs:
- T1: anatomical 3D image
Isotropic resolution recommended
Must be labeled as 'T1' modality
Minimum input requirements:
- Image FOV must contain whole skull (not be brain extracted).
- For optimal result reliability, isotropic resolution is recommended
- Recommended resolution: 1 mm isotropic.
- Minimum reliable resolution: 2mm isotropic.
Settings:
- NIfTI converter
- Preferred DICOM to NIfTI conversion tool (drop-down selection)
The selected tool will be tried first to convert DICOM to NIfTI. If the conversion fails, the other options will be tried sequentially until a successful conversion.- Mrtrix (Default)
- bru2nii
- DCM2nii
- mcverter
- diffunpack
- Preferred DICOM to NIfTI conversion tool (drop-down selection)
- Resampling
- Isotropic resolution (mm) (decimal) (Default: 1)
Choose the desired output isotropic resolution in mm. A 0 value will resample to the highest resolution dimension of the input data.
- Isotropic resolution (mm) (decimal) (Default: 1)
- Registration
- Select tool for image registrations (drop-down selection)
- ANTs (Default)
- FSL
- AC-PC alignment (checkbox)
If checked the image is AC-PC aligned (Default checked).
- Select tool for image registrations (drop-down selection)
- Skull-Stripping
- T1 Skull-stripping tool (drop-down selection)
- ROBEX (Default)
- optiBET
- ROptibeX (the addition of both ROBEX and Optibet masks)
- AFNI
- BET
- Brainsuite
- Additional skull-stripping parameters (string)
String with additional command parameters for the skull stripping tool. Optional (Default empty). - Tissue Segmentation
- Select tool for segmentation (drop-down selection)
- ANTs (Default)
- FreeSurfer v5.3.0
- Atropos (ANTs) segmentation parameters (string)
String with additional command parameters for the skull stripping tool. Optional (Default "-m 2 -n 20 -w 0.25 ").
- Select tool for segmentation (drop-down selection)
- Cortical Thickness
- Cortical thickness (checkbox)
If checked computes cortical thickness measures. Significantly increases computation time (Default checked).
- Cortical thickness (checkbox)
Outputs:
- Report:
- report.pdf: report file with results summary.
- Images:
- T1_acpc.nii.gz: preprocessed T1 image acpc-aligned.
- T1strip.nii.gz: preprocessed and skull-stripped T1 image.
- T1mask.nii.gz: binary brain-extraction mask for the T1 image.
- T1_act.nii.gz: image containing the different segmented tissues as separate volumes.
- tissueSegmentation.nii.gz: 5-tissue (csf, gm, wm, brainstem, cerebellum) tissue segmentation image.
- atlas_registered.nii.gz: template atlas labels registered into subject space.<
- full_labeled.nii.gz: parcellated labels image, using all labels provided by the selected atlas.
- labeled.nii.gz: label image with a selection of labels from of 85 relevant gray matter regions.
- thickness.nii.gz: cortical thickness values image (not present if thickness computation is de-selected).
- Datasheets:
- tissueSegmentation.csv: comma-separated-value (CSV) file with tissue types volume information How to interpret volumetry .csv outputs .
- volumetric.csv: CSV file with gray matter parcellated regions volume and cortical thickness information (includes tissue types) How to interpret volumetry .csv outputs .
Typical execution time:
- 4 hours (with cortical thickness computation).
- 1 hour (without cortical thickness computation)
Additional details:
- Cortical thickness computation is the most time-consuming step of this tool, if not required, select to skip this step and the tool will be completed in a fraction of the time.
Create free account now!
References:
- Skull-stripping:
- ROBEX skull-stripping: Iglesias et al. 2014
- optiBET skull-stripping: [Lutkenhoff et al. 2014]
- BET skull-stripping: [Smith et al. 2002]
- AFNI: [Cox 1996]
- BrainSuite skull-stripping: [Dogdas et al. 2005]
- FSL tools: [Smith et al. 2004]
- Atropos tissue segmentation: Avants et al. 2011
- ANTs registration: [Avants et al. 2008]
- DiRECT cortical thickness: Das et al. 2009
- LPBA40 Atlas: [Shattuck et al. 2008]
- Neurodevelopmental Atlases: [Almli et al. 2007], [Sanchez et al. 2011], [Sanchez et al. 2012], [Richards and Xie 2015], [Richards et al. 2015], [Fillmore et al. 2015], [Evans 2006]
- MNI152 template: Grabner et al. 2006
- Refer to ANTs / FreeSurfer for relevant references when using the respective option.
