dMRI Tractography
Generates a whole-brain set of tractography streamlines, allowing to reconstruct in 3D the main fiber pathways or most likely neural tracts constituting the white matter and connecting gray matter regions Basser et al. 2000.
The diffusion MRI information is translated into fiber orientation information using constrained spherical deconvolution (CSD) models (for HARDI data) Tournier et al. 2014 or tensor models (for DTI data) Veraart et al. 2013 .
Both probabilistic Tournier et al. 2010, Jones 2008 and deterministic tractography algorithms Tournier et al. 2012, Basser et al. 2000 are available.
Input dMRI images are fully pre-processed (optional) and scalar maps are also derived. Refer to dMRI pre-processing and FA/Scalar Maps for more information.
Deterministic Tractography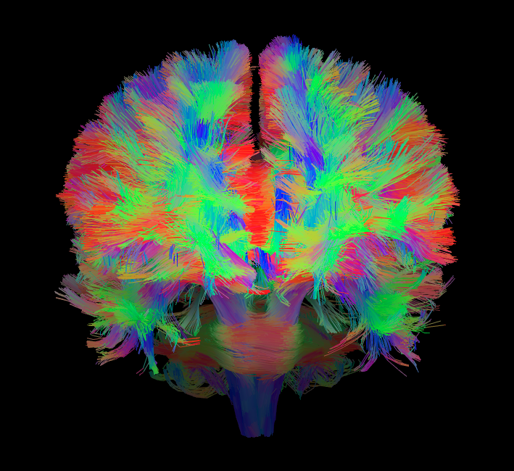
Required inputs:
- dMRI: diffusion 4D image (DTI or HARDI).
Isotropic resolution recommended.
Must be labeled as 'DTI' or 'HARDI' modality. - Gradients: gradient table (optional if dMRI is DICOM).
MRtrix format supported: file must have '.b' extension and be labelled as 'gradient_table'.
FSL format supported: files must have '.bvec' and '.bval' extensions and be labeled 'bvec' and 'bval'.
Optional inputs:
- GFMs: Gradient field-maps, recommended (magnitude image and phase image).
Same resolution and FOV as dMRI image recommended.
Files must be labeled 'gfm_magnitude' and 'gfm_phase' respectively.
Files must additionally be labelled 'dti' or 'hardi' depending on dMRI modality label.
Minimum input requirements:
- Image must contain full brain DWI dataset.
- For optimal result reliability, dMRI isotropic resolution is highly recommended
- Recommended resolution: 2 mm isotropic.
- Minimum reliable resolution: 3 mm isotropic.
- If DTI image must have at least 7 volumes (1 b0 and 6 diffusion volumes).
- If HARDI image must have at least 21 volumes (1 b0 and 20 diffusion volumes).
- For optimal preprocessing reliability, 1 b0 image is recommended every 10 gradient volumes.
Settings:
- DICOM conversion
- Preferred DICOM to NIfTI conversion tool (drop-down selection)
The selected tool will be tried first to convert DICOM to NIfTI. If the conversion fails, the other options will be tried sequentially until a successful conversion.- MRtrix (Default)
- DCM2nii
- mcverter
- diffunpack
- Preferred DICOM to NIfTI conversion tool (drop-down selection)
- dMRI pre-processing: The following information about the sequence protocol is required for optimal pre-processing. If 'Use manual diffusion acquisition parameters' is not checked, we will try to retrieve them from the DICOM header. We only provide this service for Siemens and Philips vendors. If unable to recover the true acquisition settings, we will continue with the following default settings: Echo spacing = 0.0003 s; Echo factor = 100; Acceleration factor = 1. They will work well on most datasets.
Echo Spacing (s): The time between echoes in an echo planar image (EPI).
Echo Factor: also called Echo Train Length (ETL), the number of echoes acquired in a single repetition time (TR),
Acceleration factor: the number of EPI lines acquired in parallel.
Echo Difference (s): (only needed if GFMs are provided), the difference in Echo Time (ET) between GFM acquisitions. Default value is typical for SIEMENS scanners.
- Skip pre-processing (checkbox)
If checked uses the parameters input manually in the followin - Use manual diffusion acquisition parameters (checkbox)
If checked uses the parameters input manually in the following settings (Default unchecked). - Echo Spacing (s) (float)
(Default 0.3 ms) - Echo Factor (integer)
(Default 100) - Acceleration Factor (integer)
(Default 1) - Echo Difference (s) (float)
(Default 2.46 ms) - dMRI isotropic resampling resolution (mm) (float)
Resamples the dMRI image to the indicated isotropic resolution in mm. (Default 2 mm)
- Skip pre-processing (checkbox)
- Alignment
- Do ACPC alignment (checkbox)If checked do ACPC alignment (Default checked)
- Select tool for ACPC alignment (drop-down selection)
- ANTs (default)
- FSL
- Skull-stripping
- Skull-stripping tool (drop-down selection)
- MRtrix (Default)
- AFNI
- BET
- Additional skull-stripping parameters (string)
String with additional command parameters for the skull stripping tool. Optional (Default empty).
- Skull-stripping tool (drop-down selection)
- Track generation
- Max. spherical harmonic order (only for HARDI) (single choice)
Defines the max complexity of the spherical harmonic- 4
- 6 (default)
- 8
- 10
- Tracking algorithm type (multiple choice)
Type of tractography algorithm used (Default Probabilistic)- Probabilistic (Default)
- Deterministic
- Number of streamlines (integer)
Total number of streamlines that will be computed (Default 100.000). - Step size (mm) [0 for algorithm default] (float)
The distance between consecutive calculated streamline points (Default 0).
If left at 0, a default value will be assigned depending on the type of algorithm chosen.
HARDI probabilistic: 0.5 voxelsize. Rest: 0.1 voxelsize.
- Max. spherical harmonic order (only for HARDI) (single choice)
- Track stopping criteria
- Stopping angle (º) [0 for algorithm default] (float)
The maximum angle between successive steps, default is 90deg * stepsize / voxelsize. - Min. streamline length (mm) (float)
Minimum size of a streamline to be considered as valid (Default 0). - Min. FA/FOD cutoff (float)
Minimum FA value or peak FOD value in a voxel to continue tracking (Default 0.2).
- Stopping angle (º) [0 for algorithm default] (float)
- Track filtering
- Spline-point filter factor (integer)
The factor by which the number of points in each streamline will be filtered by (Default 10). - Downsampled streamlines (integer)
Final number of streamlines to be used for visualization (Default 35.000).
- Spline-point filter factor (integer)
Outputs:
- Report:
- report.pdf: report file with results summary.
- Images:
- DWI.nii.gz: input dMRI image.
- DWI_eddy_corr.nii.gz: motion + Eddy corrected dMRI image.
- DWI_corr.nii.gz: fully corrected dMRI image (including distortion corrected if selected).
- DWI_resampled_acpc.nii.gz: final corrected, resampled and aligned dMRI image
- DWI_strip_bin.nii.gz: brain mask derived from dMRI data
- gradient_table_corr_acpc.b: gradient table corrected for AC-PC alignment and Eddy correction.
- average_b0s.nii.gz: b0 images extracted and averaged.
- FA.nii.gz: fractional anisotropy image
- AD.nii.gz, MD.nii.gz, RD.nii.gz, Cl.nii.gz, Cs.nii.gz, Cp.nii.gz: scalar maps.
- DT.nii.gz: components of the diffusion tensor fitted to the dMRI data.
- vector.nii.gz: components of diffusion tensor principal direction.
- fod.nii.gz: fiber orientation distribution file (if dataset is DTI it will be a tensor file)
- Streamlines:
- streamline.tck: full set of streamlines generated.
- streamline_downsampled.trk: a downsampled set of streamlines for 3D visualization.
Typical execution time:
- 3 hours.
References:
-
MRtrix tools: Tournier et al. 2012
- Diffusion model:
- Tensor: Veraart et al. 2013
- Constrained Spherical Deconvolution: Tournier et al. 2014
- Tracking algorithm:
- Probabilistic with HARDI data: Tournier et al. 2010
- Probabilistic with DTI data: Jones 2008
- Deterministic with HARDI data: Tournier et al. 2012
- Deterministic with DTI data: Basser et al. 2000
- Refer to dMRI pre-processing and FA/Scalar Maps for additional relevant references.
Create free account now!
