dMRI FA/Scalar Maps
Derives a fractional anisotropy (FA) map from diffusion MRI images Basser and Pierpaoli 1996 as well as maps from additional scalar measures O'Donnell and Westin 2012 : Mean Diffusivity (MD), Axial Diffusivity (AD), Radial Diffusivity (RD), Linear Component (Cl), Planar Component (Cp) and Spherical Component (Cs). It also produces tensor and eigenvector outputs derived from the dMRI image.
Works with both DTI and HARDI input images.
Input dMRI images are fully pre-processed (optional) before scalar map derivation. Refer to dMRI pre-processing for more information.
FA/Scalar Maps output example
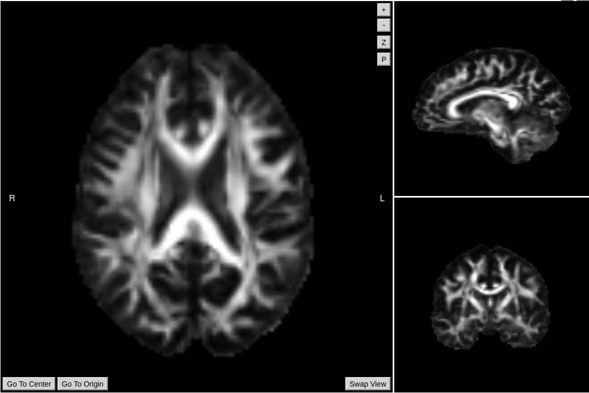
Required inputs:
- dMRI: diffusion 4D image (DTI or HARDI).
Isotropic resolution recommended.
Must be labelled as 'DTI' or 'HARDI' modality. - Gradients: gradient table (optional if dMRI is DICOM).
MRtrix format supported: file must have '.b' extension and be labelled as 'gradient_table'.
FSL format supported: files must have '.bvec' and '.bval' extensions and be labeled 'bvec' and 'bval'.
Optional inputs:
- GFMs: Gradient field-maps, recommended (magnitude image and phase image).
Same resolution and FOV as dMRI image recommended.
Files must be labelled 'gfm_magnitude' and 'gfm_phase' respectively.
Minimum input requirements:
- Image must contain full brain DWI dataset.
- For optimal result reliability, dMRI isotropic resolution is highly recommended
- Recommended resolution: 2 mm isotropic.
- Minimum reliable resolution: 3 mm isotropic.
- For DTI image must have at least 7 volumes (1 b0 and 6 diffusion volumes).
- For HARDI image must have at least 21 volumes (1 b0 and 20 diffusion volumes).
- For optimal preprocessing reliability, 1 b0 image is recommended every 10 gradient volumes.
Advanced Settings
DCM2NII
- Preferred DICOM to NIfTI conversion tool.
- Options: dcm2niix (Default), MRtrix, dcm2nii, diffunpack, mcverter.
PREPROCESSING
- Do denoising. (Default checked)
- Do Gibb ringing correction. (Default checked)
- Do Eddy. (Default checked)
- Do Fugue. (Default checked)
- Do denoising. (Default checked)
- Use manual diffusion acquisition parameters. (Default unchecked)
- If checked uses the parameters input manually in the following settings
- Echo Spacing (s). (Default 0.0003)
- The time between echoes in an echo planar image (EPI).
- Echo Factor. (Default 100)
- Also called Echo Train Length (ETL), the number of echoes acquired in a single repetition time (TR)
- Acceleration Factor. (Default 1)
- The number of EPI lines acquired in parallel.
- Echo difference (Default 0.00246)
- (only needed if GFMs are provided) The difference in Echo Time (ET) between GFM acquisitions. The default value is typical for SIEMENS scanners.
- AC-PC alignment (Default checked)
- Do bias field correction (Default checked)
- Resample dMRI image (Default checked)
- dMRI isotropic resampling resolution (mm)
- Resamples the dMRI image to the indicated isotropic resolution in mm. (Default 2 mm)
FA
- FSL Tensor. (Default unchecked)
- If tensors were computed using FSL, check this box.
Outputs:
- Parent/FA:
- report.pdf: report file with results summary.
- FA.nii.gz: fractional anisotropy image
- AD.nii.gz, MD.nii.gz, RD.nii.gz, Cl.nii.gz, Cs.nii.gz, Cp.nii.gz: scalar maps.
- vector.nii.gz: components of diffusion tensor principal direction.
- Measures:
- DT.nii.gz: components of the diffusion tensor fitted to the dMRI data.
- average_b0s.nii.gz: b0 images extracted and averaged.
- mask.nii.gz: brain mask.
- Preprocessing:
- DWI_acpc.nii.gz: final corrected, resampled and aligned dMRI image
- DWI_eddy_corr_acpc.b: gradient table corrected for AC-PC alignment and Eddy correction.
- average_b0s.nii.gz: b0 images extracted and averaged.
- diffusion_pis_voxels·nii.gz: physically implausible signal mask.
Typical execution time:
- 15 minutes.
References:
- MRtrix tools: Tournier et al. 2012
- Tensor estimation: [Veraart et al. 2013]
- Scalar measures: [Basser et al. 1994], [Westin et al. 1997] ("Geometrical diffusion measures for MRI from tensor basis analysis". Proc Intl Soc Mag Reson Med).
- Refer to dMRI pre-processing for additional relevant references.
Create free account now!
