The ANTs workflow does a tissue segmentation and a cortical thickness analysis. Another step of this workflow performs an atlas-based cortical parcellation and derives the volume of each region, followed by the computation of the average and standard deviation of each region's thickness.
The automated, volume-based Advanced Normalization Tools (ANTs) cortical thickness pipeline comprising well-vetted components such as:
- SyGN (multivariate template construction)
- SyN (image registration)
- N4 (bias correction)
- Atropos (n-tissue segmentation)
- DiReCT (cortical thickness estimation)
Click here to access the reference paper.
The full workflow includes:
- DICOM files to NIfTI conversion and reorientation to standard template images (MNI152).
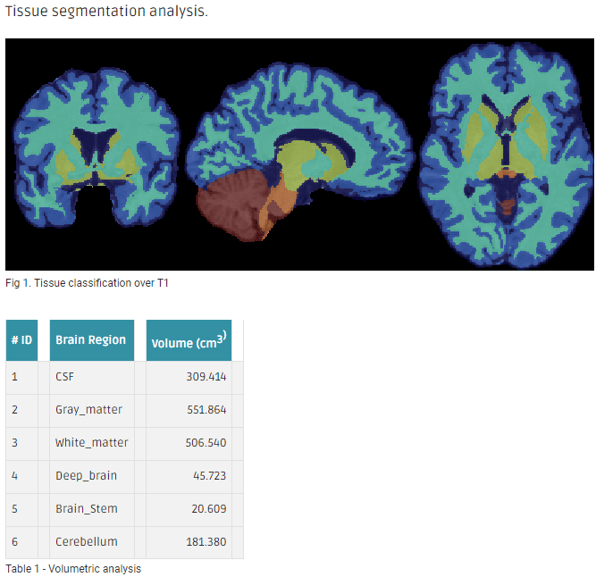
- ANTs cortical volumetry over the structural T1w image to extract the tissue segmentation file. Cortical thickness is optional.
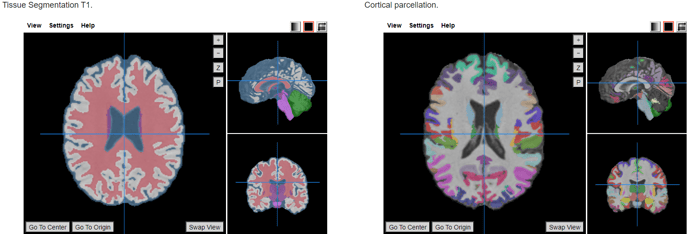
- Applies an atlas to the segmented mask of the cortex (gray matter + subcortical gray matter + brainstem + cerebellum).
Required inputs:
- T1: anatomical 3D image
- Isotropic resolution recommended
- Must be labeled as 'T1' modality
Minimum input requirements:
- For optimal result reliability, an isotropic resolution is highly recommended
- T1 recommended resolution: 1 mm isotropic.
- T1 minimum reliable resolution: 2mm isotropic.
Settings
ACPC_t1:
- Check when data is skull stripped (checkbox): Enable this if the input data is skull stripped (default unchecked)
- Change image resolution (checkbox): Changes the voxel size to isotropic resolution, defined in the next setting (default checked)
- Isotropic voxel final size (mm) (integer): Define new isotropic resolution for the T1 image (default: 1)
- ACPC alignment and remove subject face (checkbox): enable defacing step
- Output transformation mat (checkbox): get the transformation matrix in the output container.
DCM2NII:
- Preferred DICOM to NIfTI conversion tool (drop-down selection): The selected tool will be tried first to convert DICOM to NIfTI. If the conversion fails, the other options will be tried sequentially until a successful conversion.
- DCM2niix (Default)
- Mrtrix
- DCM2nii
- diffunpack
- MRIConvert (mcverter)
- Reorient images to standard space (checkbox)
ANTS
- Compute thickness map (checkbox): Perform additional cortical thickness on T1 (default unchecked)
ATLAS
- Brain atlas for parcellation (drop-down selection):
- DKT40 (Mindboggle-101) (default)
Metadata results
This workflow is not adding results as metadata.
This tool provides the volume of 89 brain regions.
References
- Avants BB, Epstein CL, Grossman M, Gee JC. Symmetric diffeomorphic image registration with cross-correlation: evaluating automated labeling of elderly and neurodegenerative brain. Med Image Anal. 2008 Feb;12(1):26-41. Epub 2007 Jun 23.
- Avants BB, Tustison NJ, Song G, Cook PA, Klein A, Gee JC., A reproducible evaluation of ANTs similarity metric performance in brain image registration. Neuroimage. 2011 Feb 1;54(3):2033-44. doi: 10.1016/j.neuroimage.2010.09.025. Epub 2010 Sep 17.
- Avants B, Duda JT, Kim J, Zhang H, Pluta J, Gee JC, Whyte J. Multivariate analysis of structural and diffusion imaging in traumatic brain injury. Acad Radiol. 2008 Nov;15(11):1360-75. doi: 10.1016/j.acra.2008.07.007.
- Tustison NJ, Avants BB, Cook PA, Kim J, Whyte J, Gee JC, Stone JR. Logical circularity in voxel-based analysis: normalization strategy may induce statistical bias. Hum Brain Mapp. 2014 Mar;35(3):745-59. doi: 10.1002/hbm.22211. Epub 2012 Nov 14.
- https://mindboggle.info/data.html
Create free account now!

