Which data formats are supported?
Supported data formats
DICOM data
Supported TAGS
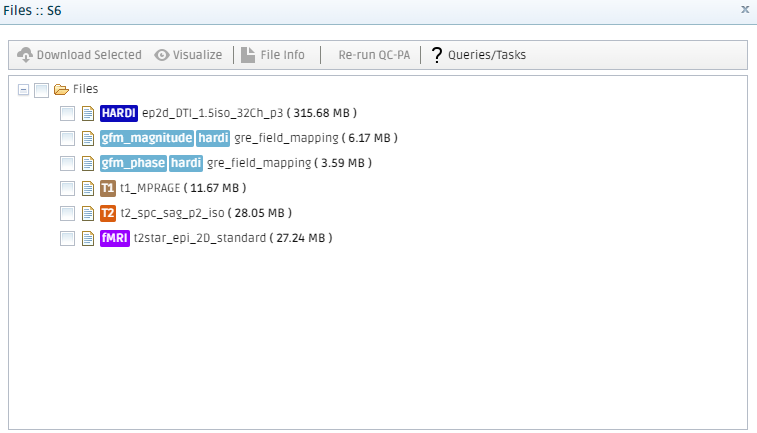
In the Subject profile tab, you can See Uploaded Files for that time point. All different sequences will be listed, and the colourful tags show what the sequence type (T1, T2, DTI, HARDI, DSI, ...). In small letters are special tags that tell the platform that those files have a specific functionality, for instance, they are gradient tables for a diffusion acquisition, gfm_magnitude, and gfm_phase for GFM preprocessing, etc ...
- "Gradient_table"
Automatically set if there’s a file with X Y Z bval, with extension .b)
- "bvec", "bval"
fsl format files, automatically set if there’s an upload file with extension .bval and .bvec
- "gfm_magnitude", "gfm_phase"
tags to set for GFM preprocessing
- "labels"
if a file has the tag, it’s nifti in the same space as the b0. Used for connectome, without having a T1. work in progress.
- "mask" (reg_mask), "mask_ref" (reg_mask)
Registers the mask image (typically a tumor) into the connectome space (the b0). Necessary for Connectome Tumor analysis.
- "roi" (stealth)
Takes a mask (for example fMRI activation map), to filter the tracts of a connectome.
Necessary for Connectome Neuronavigation analysis.
Create free account now!
