SIENAX is a package for single-time-point ("cross-sectional") analysis of brain change, in particular, the estimation of atrophy (volumetric loss of brain tissue) Smith et al. 200 . SIENAX has been used in many clinical studies.
SIENAX estimates total brain tissue volume, from a single image, normalized for skull size. It calls a series of FSL programs Smith et al. 2004 : It first strips non-brain tissue, and then uses the brain and skull images to estimate the scaling between the subject's image and standard space. It then runs tissue segmentation to estimate the volume of brain tissue, and multiplies this by the estimated scaling factor, to reduce head-size-related variability between subjects. More information on the SIENAX tool
SIENAX output examples
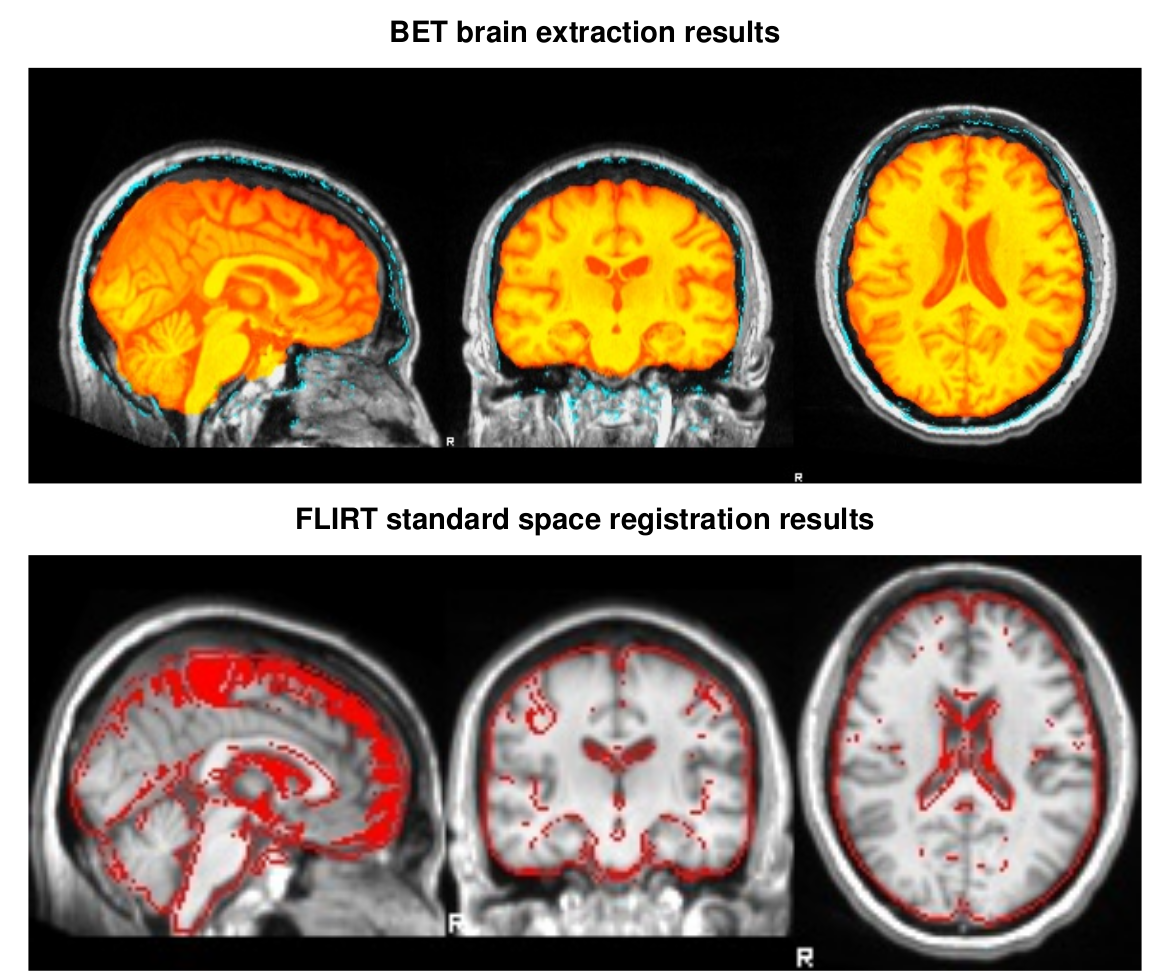
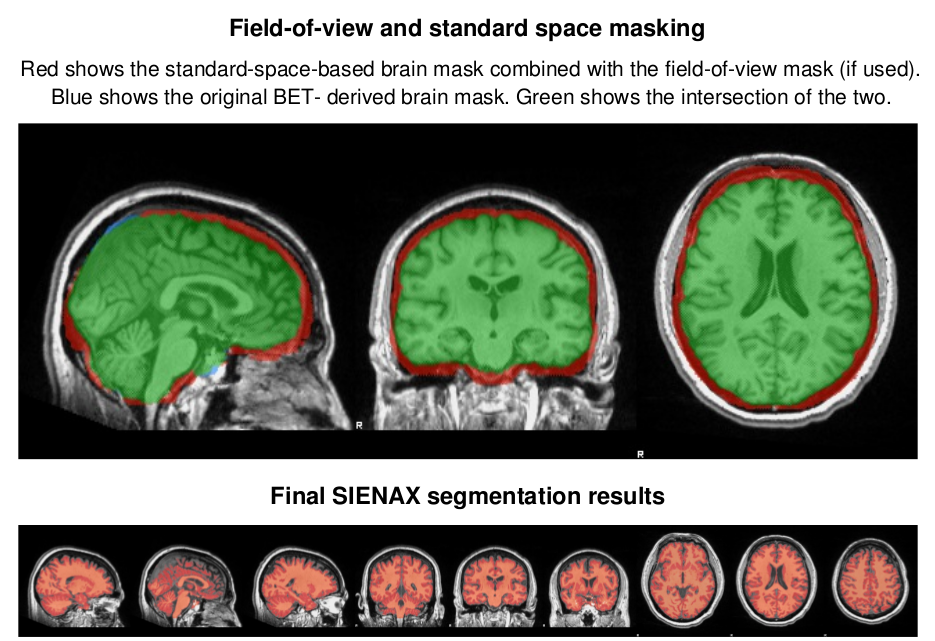
The full Workflow includes
- DICOM to NIfTI file conversion
- SIENAX 2.6
Required inputs:
- T1: anatomical 3D image
Isotropic resolution recommended
Must be labeled as 'T1' modality
Minimum input requirements:
- Image must contain full skull (not be brain extracted).
For optimal result reliability, isotropic resolution is recommended- Recommended resolution: 1 mm isotropic.
- Minimum reliable resolution: 2mm isotropic.
Outputs:
- Report:
- report.pdf: report file with results summary.
- Images:
- T1_original.nii.gz: the input T1 image used in the tool.
- T1_acpc.nii.gz: preprocessed T1 image acpc-aligned.
- Folders:
- sienax_out: Folder with full outputs of the SIENAX command.
Typical execution time:
- 15 min
References:
- SIENAX: Smith et al. 2002
- FSL tools: Smith et al. 2004
Create free account now!

