SIENA (2.6 / FSL 6.0)
SIENA is a package for two-time-point ("longitudinal") analysis of brain change, in particular, the estimation of atrophy (volumetric loss of brain tissue). SIENA has been used in many clinical studies.
Siena estimates percentage brain volume change (PBVC) betweem two input images, taken of the same subject, at different points in time. It calls a series of FSL programs to strip the non-brain tissue from the two images, register the two brains (under the constraint that the skulls are used to hold the scaling constant during the registration) and analyse the brain change between the two time points. It is also possible to project the voxelwise atrophy measures into standard space in a way that allows for multi-subject voxelwise statistical testing. An extension for ventricular analysis is provided in FSL5.
SIENA output examples
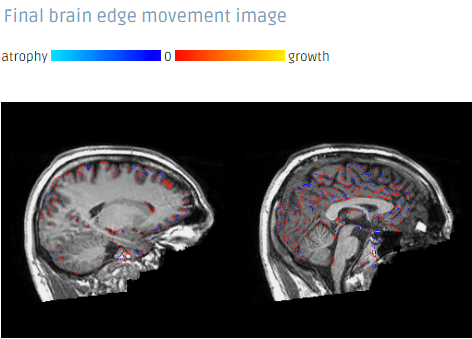
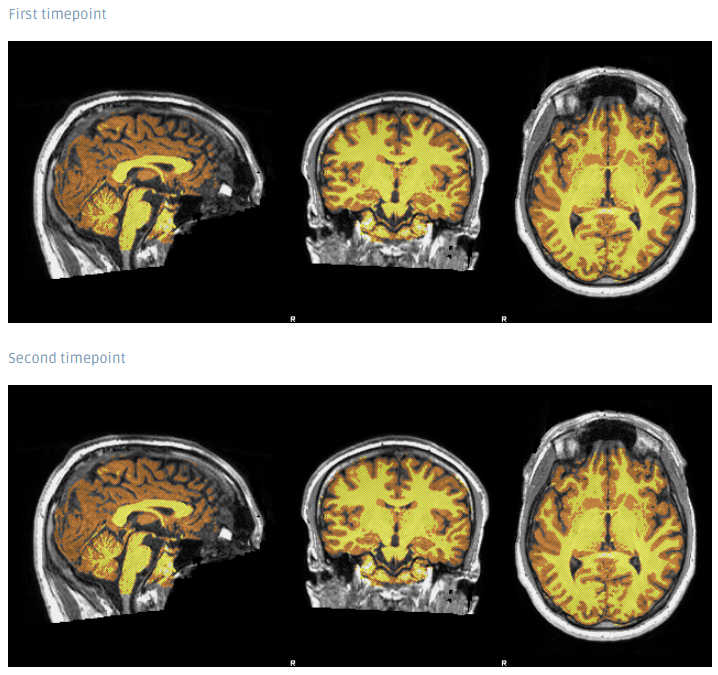
The full Workflow includes
- DICOM to NIfTI file conversion
- SIENA
Required inputs
- T1 (Timepoint 1): Anatomical 3D image
- Image must contain full skull (not be brain extracted).
- Must be labeled as T1 modality
- T1 (Timepoint 2): Anatomical 3D image
- Image must contain full skull (not be brain extracted).
- Must be labeled as T1 modality
Minimum input requirements
- For optimal result reliability, isotropic resolution is recommended
- Recommended resolution: 1 mm isotropic.
- Minimum reliable resolution: 2mm isotropic.
Output files
- Report: Structured report in pdf format including the most relevant results of the analysis.
- SIENA files: All the output files from SIENA.
Typical execution time
- 30 minutes
References
- SIENA: Smith et al. 2002
- FSL tools: Smith et al. 2004
Create free account now!
