SAMSEG MS Lesion Segmentation Workflow w/ Editor
This workflow uses FreeSurfer’s SAMSEG tool to robustly segment 41 brain structures and white matter lesions in multiple sclerosis using an algorithm adaptive to different scanners and input sequences without the need for retraining. The workflow accepts up to five different input contrasts with minimal preprocessing, and performs automatic segmentation before offering the user the possibility of manual lesion mask editing for the correction of false positives/negatives. It is suitable for use with both MS and control brains.
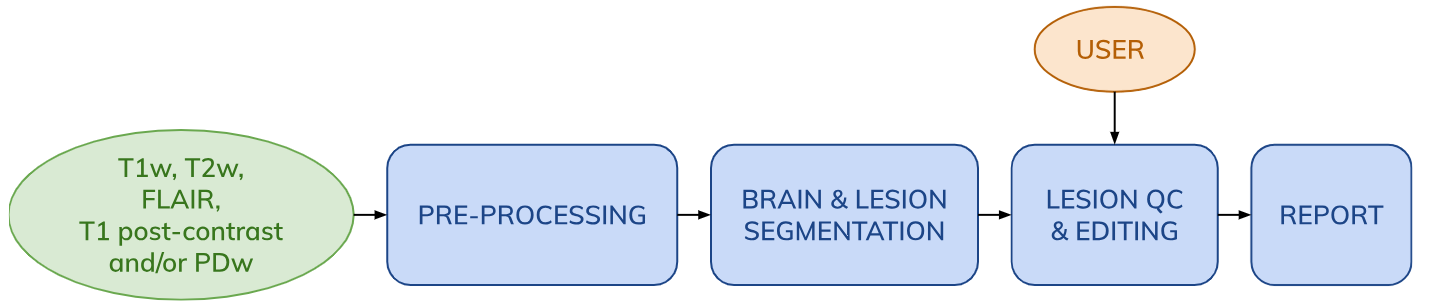
The workflow includes the following steps:
- Image pre-processing including DICOM to NIfTI conversion (if applicable) and registration of all inputs to the contrast with the highest resolution.
- SAMSEG automatic whole-brain and MS lesion segmentation.
- Semi-automatic lesion mask QC and editing using the QMENTA Viewer.
- PDF report compiling visuals of brain and lesion segmentation results, as well as lesion statistics such as lesion counts and volumes.
Note: Image pre-processing in the SAMSEG workflow is minimal - reformatting to 1mm isotropic resolution, bias field correction and skull stripping are neither needed nor recommended.
Workflow Scheme
SAMSEG Workflow output example
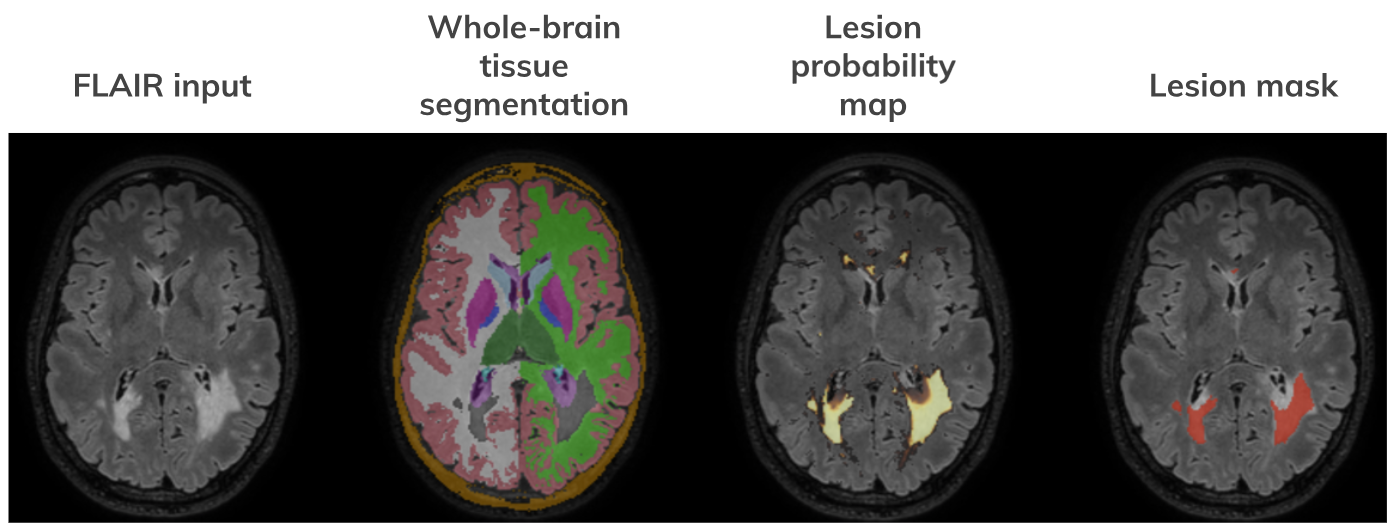
Note: The correspondence between structures segmented and their neuroanatomical labels is according to $FREESURFER_HOME/FreeSurferColorLUT.txt. In the whole-brain tissue segmentation, lesions are assigned a value of 99.
Lesion editing with the QMENTA viewer
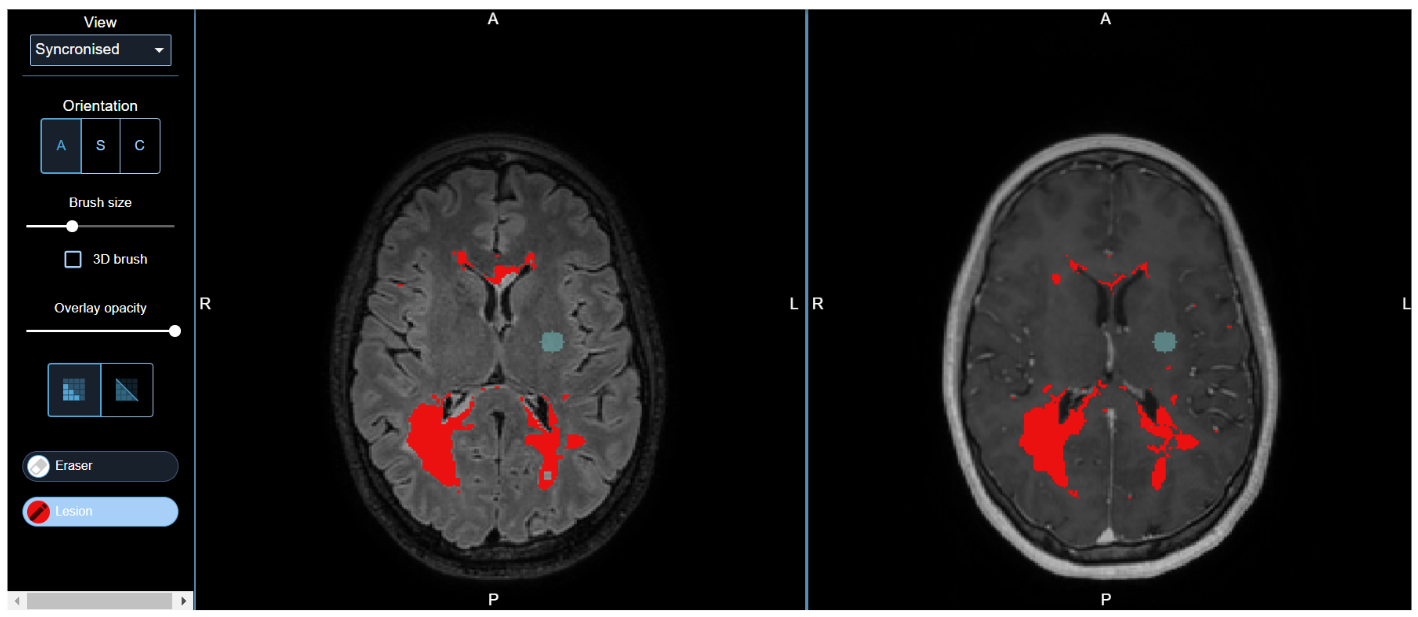
See QMENTA Label/Segmentation Editor for more information.
Required inputs:
The following input sequences are accepted, one of each and up to a total of five:
- T1: 3D anatomical T1-weighted image. Must be labeled with 'T1' modality.
- T2: 3D anatomical T2-weighted image. Must be labeled with 'T2' modality.
- FLAIR: 3D T2-FLAIR image. Must be labeled with 'T2' modality and 'flair' tag.
- T1 post-contrast: 3D T1-weighted post-contrast (gadolinium) image. Must be labeled with 'T1' modality and ‘post_contrast’ tag.
- PD: 3D proton density-weighted image. Must be labelled with 'PD' modality.
Note: For optimal lesion segmentation results, a T1w-FLAIR combination is recommended.
Minimum input requirements:
- Whole-brain FOV
- Recommended resolution: 1 mm isotropic.
Workflow settings:
- Segment pallidum (checkbox). Controls whether to treat the pallidum internally as a white matter structure, or to segment it separately. Only recommended with the inclusion of input contrasts where the pallidum is clearly discernible (e.g. T2w or FLAIR).
- Intensity masking (checkbox). By default, lesion voxels are masked based on the mean intensity of the grey matter tissue class. You can relax this by masking lesion voxels using white matter instead. But be aware, these changes might lead to an increase in false positive lesions.
- Lesion probability map threshold (default 0.3). Controls the probability threshold at which a particular voxel is considered to belong to a lesion, after having introduced lesion shape constraints. The lesion mask output is initialized by binarizing the posteriors map using this threshold.
- Editor base volumes (checkboxes). If > 2 input contrasts are used, select two of these to display as base volumes during editing of the lesion mask. If none are selected, volumes will be chosen automatically based on their intensity contrast profiles.
Note: MS lesion segmentation is enabled by default in this workflow. If you would like to explore SAMSEG’s basic functionality of whole-brain segmentation alone, please get in touch.
References:
FreeSurfer Wiki: https://surfer.nmr.mgh.harvard.edu/fswiki/Samseg
Cerri, S., Puonti, O., Meier, D. S., Wuerfel, J., Mühlau, M., Siebner, H. R., & Van Leemput, K. (2021). A contrast-adaptive method for simultaneous whole-brain and lesion segmentation in multiple sclerosis. NeuroImage, 225, 117471. https://doi.org/10.1016/j.neuroimage.2020.117471
Puonti, O., Iglesias, J. E., & Van Leemput, K. (2016). Fast and sequence-adaptive whole-brain segmentation using parametric Bayesian modeling. NeuroImage, 143, 235–249. https://doi.org/10.1016/j.neuroimage.2016.09.011
Create free account now!
