Q-FiberMapper
This tool uses a T1 weighted anatomical image and a DWI acquisition to extract the major fascicles of the brain, and derive diffusion metrics specific to each fascicle including fractional anisotropy (FA), mean diffusivity (MD), radial diffusivity (RD), axial diffusivity (AD) and the coefficients of planarity (Cp), linearity (Cl) and sphericity (Cs). Outputs the reports in neurological and radiological orientations.
For more information, please check the tool manual here.
The QFM reconstructs the following 16 fascicles:
- Left and right inferior fronto-occipital fasciculus (IFOF)
- Left and right inferior longitudinal fasciculus (ILF)
- Left and right superior longitudinal fasciculus (SLF)
- Left and right cortico spinal tract (CST)
- Left and right uncinate fasciculus (UNC)
- Left and right cingulum bundle
- Left and right fornix
- Forceps major
- Forceps minor
For each fascicle, 7 diffusion parameters are derived and the mean and standard deviation over the volume of the fascicle listed in the table below the visualization of the reconstructed fascicle. The diffusion parameters are derived from the diffusion tensor, a mathematical model that describes the mobility of water within the tissue
Fractional anisotropy (FA): a unitless parameter that describes the directional bias of the water diffusion pattern. FA scales between 0 and 1. A value near 1 indicates that the diffusion pattern is highly directionally biased such as in deep white matter, a value near 0 indicates that the diffusion pattern is more spherical, with water diffusing equally in every direction.
Mean diffusivity (MD): this parameter describes the mean rate of diffusion in every direction, measured in 10-3 mm2/s
Axial diffusivity (AD): this parameter describes the rate of diffusion along the axis of the white matter fibers, measured in 10-3 mm2/s
Radial diffusivity (RD): this parameter describes the rate of diffusion perpendicular to the axis of the white matter fibers, measured in 10-3 mm2/s
Coefficients of linearity (Cl), planarity (Cp) and sphericity (Cs): these unitless parameters describe the linearity, planarity and sphericity of the water diffusion pattern respectively.
When tissue changes occur at a cellular level, such as the breakdown of various cellular structures the diffusion pattern of water within the tissue changes. This will be reflected in the diffusion parameters FA, MD, AD and RD. Generally, pathological tissue changes tend to increase the mobility of water within the tissue, leading to increases in MD, AD or RD. FA can increase or decrease due to pathological tissue changes. Gradual changes in all parameters are expected in healthy aging, however, extreme changes could have a pathological cause.
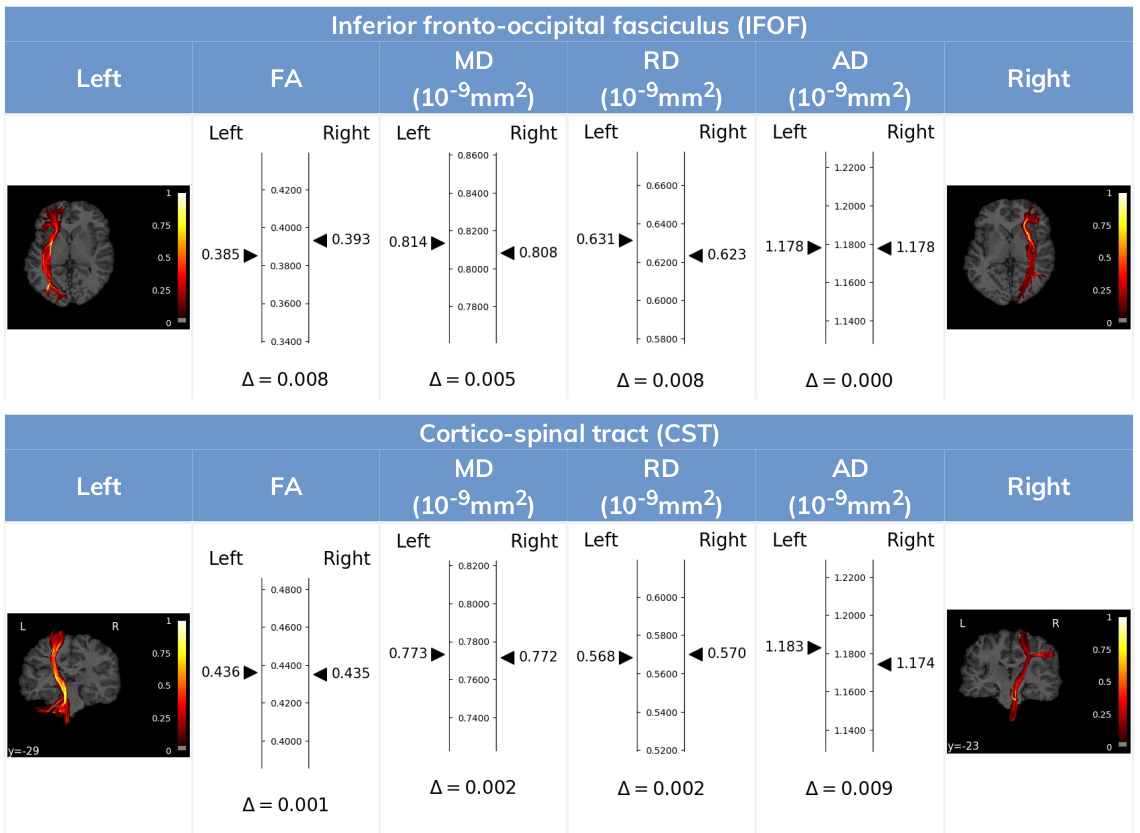
Contralateral asymmetry report
Our contralateral report provides a convenient way to compare the quantification outputs from our FiberMapper Tool of corresponding white matter fascicles of the same subject across opposing hemispheres (e.g: compare the properties of the left corticospinal tract with the right corticospinal tract). This can be useful in cases such as traumatic brain injury where trauma affects often only one of the hemispheres and changes to fascicle diffusion-derived properties can be more easily assessed using the non-affected counterpart as a baseline.
The report shows for each bilateral pair of fascicles the values corresponding to the main diffusion parameters indicated to each corresponding side of a central measuring axis. Below that axis, the absolute value of the difference between tracts is displayed.
Tract segmentation tool output example:
 |
 |
 |
 |
The full workflow includes:
-
DICOM files to NIfTI conversion and reorients to standard. The workflow file filter sends the dicom or nifti files required for the workflow to run and the box converts the dicom to niftis, extracts the necessary files (if possible) to do the dmri preprocessing step (eddy) and gradient tables. If nifti files or gradient tables are found in the input, it just does the reorient to standard step and outputs the files.
-
dMRI preprocessing includes denoising, gibbs ringing correction, eddy correction, motion correction, bias field correction, distortion correction (if gradient field maps are available) and ACPC alignment.
- Gets the brain mask and computes the tensor from the diffusion image, plus derives different scalar measures: FA, AD, MD, RD, Cl, Cs, Cp.
-
Runs ants cortical volumetry script over T1 image to extract the tissue segmentation file.
-
Applies an atlas to the segmented mask of the cortex (gray matter + subcortical gray matter + brainstem + cerebellum)
-
Align the diffusion image to the T1.
-
Estimate the diffusion model. Does nothing if the data is DTI. Runs constrained spherical deconvolution model.
-
Perform probabilistic or deterministic tractography over the diffusion model.
-
Extracts fascicles from tractography database, creates probabilistic maps of fascicle membership
- Outputs per fascicle mean diffusion metric fractional anisotropy (FA), mean/radial/axial diffusivity (MD, RD, AD), coefficient of linearity/planarity/sphericity (Cl, Cp, Cs).
-
Contralateral report comparing fascicles and their diffusion metrics with their correspondent across hemispheres.
-
Outputs PNG are presented in both orientations, radiological and neurological.
Required inputs:
- T1: anatomical 3D image
Isotropic resolution recommended
Must be labeled as 'T1' modality - dMRI: diffusion 4D image (DTI or HARDI).
Isotropic resolution recommended.
Must be labeled as 'DTI' or 'HARDI' modality. - Gradients: gradient table (optional if dMRI is DICOM).
MRtrix format supported: file must have '.b' extension and be labeled as 'gradient_table'.
FSL format supported: files must have '.bvec' and '.bval' extensions and be labeled 'bvec' and 'bval'.
Minimum input requirements:
- Image must contain full brain DWI dataset.
- For optimal result reliability, an isotropic resolution is highly recommended
- T1 recommended resolution: 1 mm isotropic.
- T1 minimum reliable resolution: 2mm isotropic.
- dMRI recommended resolution: 2 mm isotropic.
- dMRI minimum reliable resolution: 3 mm isotropic.
- T1 recommended resolution: 1 mm isotropic.
- If DTI image must have at least 7 volumes (1 b0 and 6 diffusion volumes).
- If HARDI image must have at least 21 volumes (1 b0 and 20 diffusion volumes).
- For optimal preprocessing reliability, 1 b0 image is recommended every 10 gradient volumes.
Optional inputs:
- GFMs: Gradient field-maps, recommended (magnitude image and phase image).
Same resolution and FOV as dMRI image recommended.
Files must be labeled 'gfm_magnitude' and 'gfm_phase' respectively.
Files must additionally be labeled 'dti' or 'hardi' depending on dMRI modality label.
Advanced Settings:
DCM2NII:
- Preferred DICOM to NIfTI conversion tool (drop-down selection)
Options: dcm2niix (Default), MRtrix, dcm2nii, diffunpack, mcverter. - Reorient images to standard space (checked by default)
ACPC alignment
- T1 data is skull stripped. Check this option if the input is already skull stripped. (checked by default)
- Change image resolution. Resample the data to another isotropic resolution. (checked by default)
- Isotropic voxel final size (mm). (Default value: 1)
- ACPC alignment and remove subject face. (checked by default)
PREPROCESSING
- Do denoising (checked by default)
- Do Gibb ringing correction (checked by default)
- Do Eddy (checked by default)
- Do Fugue (checked by default)
- Use manual diffusion acquisition parameters (unchecked by default): If checked, the tool will prioritize the parameters defined by the user in this menu. Echo Spacing (s) (float) The time between echoes in an echo planar image (EPI) (Default 0.0003ms)
- Echo Factor (integer)Also called Echo Train Length (ETL), the number of echoes acquired in a single repetition time (TR) (default 100)
- Acceleration Factor (integer) The number of EPI lines acquired in parallel (Default 1)
- Echo Difference (s) (float) (only needed if GFMs are provided), the difference in Echo Time (ET) between GFM acquisitions. (default 0.00246 for SIEMENS acquisitions)
- AC-PC alignment (checked by default)
- Resample dMRI image (checked by default)
- dMRI isotropic resampling resolution (mm) (float) Resamples the dMRI image to the indicated isotropic resolution in mm. (Default value is 2 mm)
MASK_ALIGNMENT
- Compute non-linear registration EPI to T1 (FA input required) (checkbox) (default checked)
ATLAS
- Brain atlas for parcellation (drop-down selection) The selected tool will be tried first to convert DICOM to NIfTI. If the conversion fails, the other options will be tried sequentially until a successful conversion. DKT40 (Mindboggle-101) (default) AAL
MODEL
- Tracking algorithm type (drop-down selection)
- Constrained Spherical Deconvolution (CSD) (default)
- Tensor
- Multi-tissue constrained spherical deconvolution
TRACTO
- Max. spherical harmonic order (drop-down menu):
- 4, 6 (default), 8, 10
- Tracking algorithm type
- DET deterministic
- PROB probabilistic
- Number of streamlines (integer) Number of streamlines to generate (default 1M)
- Step size (mm) (0 for adaptative default) (float) (default 0)
- Use Anatomical constrained tractography (checkbox). Only works when Morphology results for ACT are passed as input (default checked)
- Stopping angle (degrees) (0 for adaptive default) (integer) (default 0)
- Min. streamline length (mm) (0 for adaptive default) (integer) (default 0)
- Max. streamline length (mm) (0 for adaptive default) (integer) (default 0)
- Min. FA/FOD cutoff (float) (default 0)
REPORT
- Report default orientation. Orientation of the default report.pdf.
- Neurological orientation
- Radiological orientation
Output files
We explain the output files for each box. The modalities are shown in colored boxes and the tags are shown with outlined boxes. Each element represents a relevant file from the container, more might be present but they are not as important.
DCM2NII_dmri
- T1 : T1 NIfTI file
- HARDI / DTI : dMRI NIfTI file
- HARDI / DTI:
- HARDI / DTI ACQ_PARAMS: acquisition parameter file for eddy preprocessing automatically extracted from dicom file
- HARDI / DTI BVAL: BVAL file FSL format
- HARDI / DTI BVEC: BVEC file in FSL format
- HARDI / DTI GRADIENT_TABLE: gradient table in mrtrix format
- HARDI / DTI INDEX: index file for eddy preprocessing automatically extracted from dicom file
ACPC_alignment
- T1 HEAD Full T1 head image
- T1 BRAIN Skull stripped T1
ANTS
- T1 HEAD Full T1 head image
- T1 BRAIN Skull stripped T1
- T1 MASK Binary skull stripped brain mask
- TISSUE_SEGMENTATION 6-tissue segmentation image (1: CSF, 2: gray matter, 3: white matter, 4: deep gray matter, 5: brainstem, 6: cerebellum)
- CSV Volumetry information for ICV and each segmented tissue.
PREPROCESSING
- HARDI / DTI : dMRI NIfTI file processed with the steps selected in the settings
- GRADIENT_TABLE: gradient table in mrtrix format
- AVERAGE_B0S: Average b0s from the processed dMRI image
- REPORT: PDF report of the box
MEASURES
- TENSOR: Diffusion tensor data
- AVERAGE_B0S: Average b0s from the processed dMRI image
- MASK: dMRI binary brain mask
FA
- SCALAR AD: AD scalar file
- SCALAR CL: Cl scalar file
- SCALAR CP: Cp scalar file
- SCALAR CS: Cs scalar file
- SCALAR FA: FA scalar file
- SCALAR MD: MD scalar file
- SCALAR RD: RD scalar file
- REPORT : PDF report of the box
ATLAS
- ATLAS_INFO : Text file with atlas information
- LABELS : Labeled cortical brain
- REPORT : PDF report of the box
- Vectorized data folder
- VECTOR : Volumetry information in a vector format
MASK_ALIGNMENT
- HARDI / DTI : dMRI NIfTI file aligned to the anatomical image
- SCALAR AD: AD scalar file aligned to the anatomical image
- SCALAR CL: Cl scalar file aligned to the anatomical image
- SCALAR CP: Cp scalar file aligned to the anatomical image
- SCALAR CS: Cs scalar file aligned to the anatomical image
- SCALAR FA: FA scalar file aligned to the anatomical image
- SCALAR MD: MD scalar file aligned to the anatomical image
- SCALAR RD: RD scalar file aligned to the anatomical image
- ACT : ACT volume (5tt format) for connectome
- AVERAGE_B0S: Average b0s from the processed dMRI image aligned to the anatomical image
- GRADIENT_TABLE: gradient table in mrtrix format
- LABELS : Labeled cortical brain for connectome
- MASK: dMRI binary brain mask aligned to the anatomical image
- TISSUE_SEGMENTATION 6-tissue segmentation image for connectome
MODEL
- CSD / DTI / In the multi-tissue case we have:
- MSMT WM
- MSMT GM
- MSMT CSF
- RESPONSE
TRACTO
- TRACKS : Full tractography
- TRACKS DOWNSAMPLED : Tractography for visualization
QFM
- fa/md/rd/ad/cl/cp/cs.csv: table with the scalar metrics over each tract
- fa/md/rd/ad/cl/cp/cs_asym.csv: table with the asymmetry metrics over each bilateral tract pair
- SEG.nii.gz: a labeled segmentation of all tracts
- FASCILCLE_INFO.csv: information of streamline reconstruction for each tract
- T1strip.nii.gz
- ANAT_MAP
- _tractname_.nii.gz: visitation map for each tract in T1 space
- DIFF_MAP
- _tractname_.nii.gz: visitation map for each tract in dMRI space
- IMAGE
- _tractname_.png: neurological convention png images included in the report
- _tractname_radiological.png: radiological convention png images included in the report
- TRACK
- _tractname_.trk: streamline file for each tract
REPORT
Same files as the previous and:
- SCALAR AD: AD scalar file
- SCALAR CL: Cl scalar file
- SCALAR CP: Cp scalar file
- SCALAR CS: Cs scalar file
- SCALAR FA: FA scalar file
- SCALAR MD: MD scalar file
- SCALAR RD: RD scalar file
- report.pdf (merged contralateral report + simple tracts measures): orientation defined by settings
- Neurological reports: - report_contralateral_neurological.pdf - report_tracts_neurological.pdf
- Radiological reports: - report_contralateral_radiological.pdf - report_tracts_radiological.pdf
Create free account now!
References
dMRI PREPROCESSING
-
DWI gradient table check [Ben Jeurissen et al. 2014]
- Subject motion & Eddy correction: [Andersson and Sotiropoulos 2016]
- Distortion correction: [Jezzard and Balaban 1995].
ACPC alignment:
-
ANTs registration: [Avants et al. 2008]
- MNI152 Atlas: [Grabner et al. 2006]
-
FLIRT registration: [Jenkinson et al. 2002]
dMRI tensor and scalar measures
- MRtrix tools: [Tournier et al. 2012]
- Tensor estimation: [Veraart et al. 2013]
- Scalar measures: [Basser et al. 1994], [Westin et al. 1997] ("Geometrical diffusion measures for MRI from tensor basis analysis". Proc Intl Soc Mag Reson Med).
T1 segmentation and cortical labels
-
ANTs script: [Tustison et al. 2014]
-
ANTs registration: [Avants et al. 2008]
-
Atropos tissue segmentation: [Avants et al. 2011]
-
DiRECT cortical thickness: [Das et al. 2009]
-
OASIS-30 tissue priors: [Landman and Warfield 2012] ("MICCAI 2012 workshop on multi-atlas labeling." Medical image computing and computer-assisted intervention conference)
-
DKT40 Mindboggle 101 Atlas: [Klein and Tourville, 2012]
-
AAL Atlas: [Tzourio-Mazoyer et al. 2002], [Rolls et al. 2015]
-
MNI152 Atlas: [Grabner et al. 2006]
MASK_ALIGNMENT
- ANTs registration: [Avants et al. 2008]
Diffusion MODEL
- msmt_5tt [Ben Jeurisen et al. 2014]
- CSD [J-Donald Tournier et al. 2007]
- fod2dec: Two posters [T. Dhollander et al., 2015] [T. Dhollander et al. 2015]
TRACTO
- iFOD1 or SD_STREAM: [J-Donald Tournier et al. 2012]
- iFOD2 poster [J-Donald Tournier et al. 2010]
- Tensor_Det [Basser, P.J. et al. 2000]
- Tensor_Prob [Jones, D. et al. 2008]
- act, -backtrack, -seed_gmwmi [Robert E. Smith et al. 2012]
