Lesion TOADS Workflow
Algorithm for simultaneous brain structures and MS lesion segmentation of MS Brains. The brain segmentation is topologically consistent and the algorithm can use multiple MR sequences as input data.
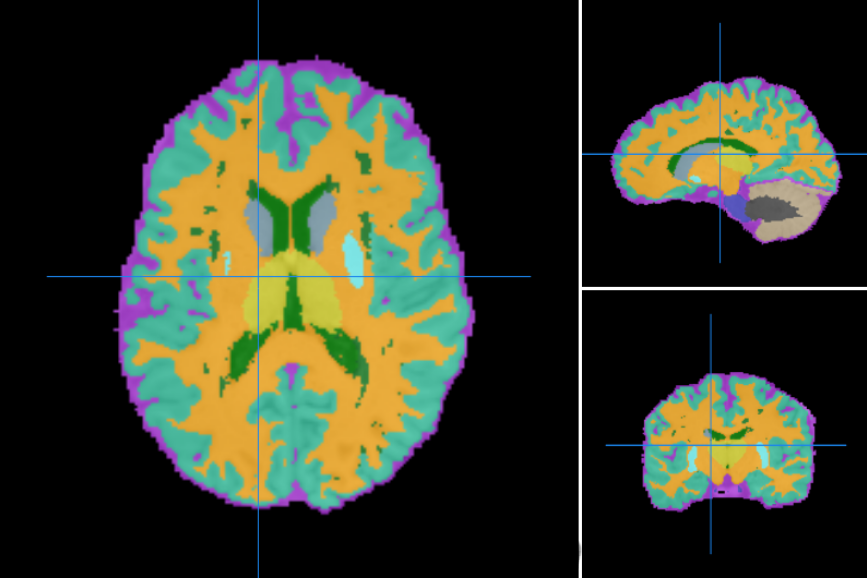
Segments brain tissues and lesions of brain images with multiple sclerosis lesions using a topology-preserving approach. It takes two anatomical images as input, a T1 from a MPRAGE or SPGR sequence and a T2 FLAIR. Both images are ACPC aligned, rigidly registered to each other and skull striped (the T1 image is additionally bias-field corrected).Then the segmentation is computed using an atlas-based technique that combines topological and statistical atlas, the resulting segmented structures are topologically constrained. It also computes the volumes for each segmented tissue in mm3.
The tool can also run with a T1 with different sequence but a positive outcome is not guaranteed. In that case, files are processed as SPGR.
The segmented structures are the following:
- Cerebrospinal fluid
- Sulcal CSF
- Ventricular CSF
- Grey matter
- Cerebral cortical gray matter
- Cerebellar cortical gray matter
- Caudate
- Putamen
- Brainstem
- White matter
- Cerebral white matter
- Cerebellar white matter
- Lesions
The full workflow includes:
-
DICOM files to NIfTI conversion and reorientation to standard.
-
AC-PC Alignment of the T1 image.
- Bias-field correction of the T1 image.
-
Skull stripping of the T1 image.
- Skull stripping of the T2 image by rigid alignment to the T1 image and applying the mask resulting from the previous step.
- Uses LesionTOADS 1.9R from NITRC to segment the tissues using the two skull stripped images.
Required inputs:
- T1-MPRAGE or T1-SPGR or T1: anatomical 3D image
Isotropic resolution recommended
Should be labelled as 'T1' modality and tagged as 'mprage' or 'spgr' depending on its sequence.
If there are no tags, the tool will process the T1 image as if it was 'spgr'
- T2-FLAIR: anatomical 3D image with hyper-intense lesions
Isotropic resolution recommended
Must be labelled as 'T2' modality and tagged as 'flair'.
Minimum input requirements:
- For optimal result reliability, an isotropic resolution is highly recommended
- T1 and T2 recommended resolution: 1 mm isotropic.
- T1 and T2 minimum reliable resolution: 2mm isotropic.
- T1 and T2 recommended resolution: 1 mm isotropic.
Settings
DCM2NII:
- Preferred DICOM to NIfTI conversion tool (drop-down selection):
The selected tool will be tried first to convert DICOM to NIfTI. If the conversion fails, the other options will be tried sequentially until a successful conversion.- DCM2niix (Default)
- Mrtrix
- DCM2nii
- diffunpack
- MRIConvert
ACPC alignment and defacing:
- Input data is skull stripped (checkbox)
- Default checked.
- Change image resolution (checkbox)
- Resamples input image to desired pixel resolution (Default checked and set to 1 mm).
- AC-PC alignment and defacing.
- Applies the alignment and the defacing based on an MNI template.
Skull Strip:
- Preferred skull stripping tool (drop-down selection):
- ROBEX (Default)
- BET
- AFNI
- Brainsuite
- MRtrix for MRI
- ANTs
- Optibet
- Optibet and ROBEX
- Apply Bias Field correction (checkbox).
- Applies N4 bias field correction to the output (Default checked).
- Additional skull-stripping parameters (string)
- String with additional command parameters for the skull stripping tool. Optional (Default empty).
- Age in months (float)
- Optional (Default 1000)
Rigid alignment and masking:
- Use transformation matrix if available (checkbox).
- The alignment can be performed using a given mat file if needed (Default unchecked).
- Keep original dimensions (checkbox).
- If selected, the reference image is resampled to the resolution of the input image (Default unchecked).
- Apply mask if available (checkbox).
- Applies a binary mask to the input image (Default checked).
- Keep image in input space (checkbox).
- If selected the image won't be aligned to the reference, can be used to mask images in input space (Default unchecked).
Lesion TOADS:
- Atlas to use (drop-down selection):
- With Lesion (Default)
- No Lesion
- Output images (drop-down selection):
- Hard segmentation
- Hard segmentation + membership
- CRUISE inputs (Default)
- Dura removal inputs
- Additional inputs (set of checkboxes):
- Hard class using max membership (Default unchecked).
- Correct MR field inhomogeneity (Default unchecked).
- Estimated inhomogeneity field (Default unchecked).
- Maximum distance from the inter-ventricular WM boundary to down-weight the lesion membership to avoid false positives (integer)
- Default 3
- Minimum ventricle distance (integer)
- Default 2
- Maximum ventricle distance (integer)
- Default 25
- Include lesion in WM class in hard classification (checkbox)
- Default checked
- Controls the effect of the statistical atlas on the segmentation (decimal)
- Default 2
- Controls the effect of neighborhood voxels on the membership (decimal)
- Default 0.2
- Maximum amount of relative change in the energy function considered as the convergence criteria (decimal)
- Default 0.0001
- Maximum iterations (integer)
- Default 40
- Atlas alignment (drop-down selection):
- Rigid
- Multi-fully-affine (Default)
- Connectivity (drop-down selection):
- (18,6) (Default)
- (6,26)
- (26,6)
- (6,18)
Output container files
We explain the output files for each box. The modalities are shown in colored boxes and the tags are shown with outlined boxes. Each element represents a relevant file from the container, more might be present but they are not as important.
DCM2NII
- T1MPRAGE/SPGR : T1-MPRAGE / T1-SPGR Nifti file
- T2FLAIR: T2-FLAIR Nifti file
ACPC alignment and defacing
- T1HEAD : T1 ACPC aligned and defaced
Skull Strip
- T1BRAIN : T1 Skull stripped
- MASK : Binary mask of the skull stripping
Rigid alignment and masking
- T2ALIGNEDMASKED : T2 aligned to T1 and masked
Lesion TOADS
- T1BRAINMPRAGE/SPGR : Input T1 image (skull stripped)
- T2FLAIR: Input T2 image (skull stripped)
- Toads_csf.nii.gz: Segmented CSF Nifti
- Toads_gm.nii.gz: Segmented gray matter Nifti
- Toads_lesions_seg.nii.gz: Segmented lesions Nifti
- Toads_seg.nii.gz: Tissue segmentation Nifti
- Toads_seg_no_background.nii.gz: Tissue segmentation Nifti with no background, used for better visualization.
- Toads_wmFill.nii.gz: Segmented white matter Nifti
- Toads_wmMask.nii.gz: Binary mask of the white matter
- REPORTCSV : csv file with the volumes of each segmented tissue in mm3
Create free account now!
References
ACPC alignment and defacing
-
ANTs registration: [Avants et al. 2008]
- MNI152 Atlas: [Grabner et al. 2006]
-
FLIRT registration: [Jenkinson et al. 2002]
Skull Strip
- ROBEX skull-stripping: Iglesias et al. 2014
- optiBET skull-stripping: [Lutkenhoff et al. 2014]
- BET skull-stripping: [Smith et al. 2002]
- AFNI: [Cox 1996]
- BrainSuite skull-stripping: [Dogdas et al. 2005]
Rigid alignment and masking
- ANTs registration: [Avants et al. 2008]
