FreeSurfer 6.0 Hippocampal Stream
Full morphological processing of a T1 anatomical image using FreeSurfer's recon-all script [Dale et al. 1999], including integration of the additional Hippocampal Subfields module. This module accepts high-resolution, dedicated hippocampal images in order to automatically segment hippocampal subfields, though can also be executed without any additional scan and using a single T1 input. More information can be found here.
Includes:
- Image pre-processing: intensity normalization, skull-stripping.
- Tissue segmentation into CSF, cortical grey matter, subcortical grey matter, white matter, brainstem and cerebellum.
- Atlas-based parcellation and labelling using DKT40 scheme.
- Cortical thickness computation for cortical regions.
- Hippocampal Subfields Segmentation.
Volumetric report output samples
Fig 1 - Volumetric information
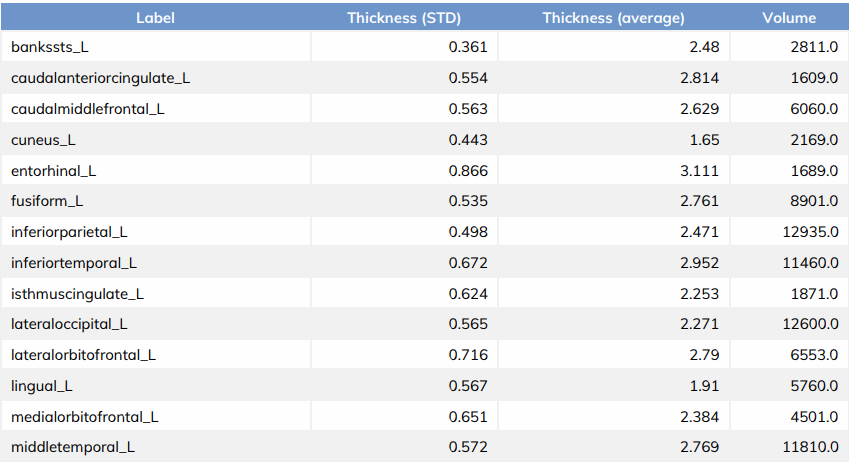
Fig 2 - Freesurfer recon-all segmentation
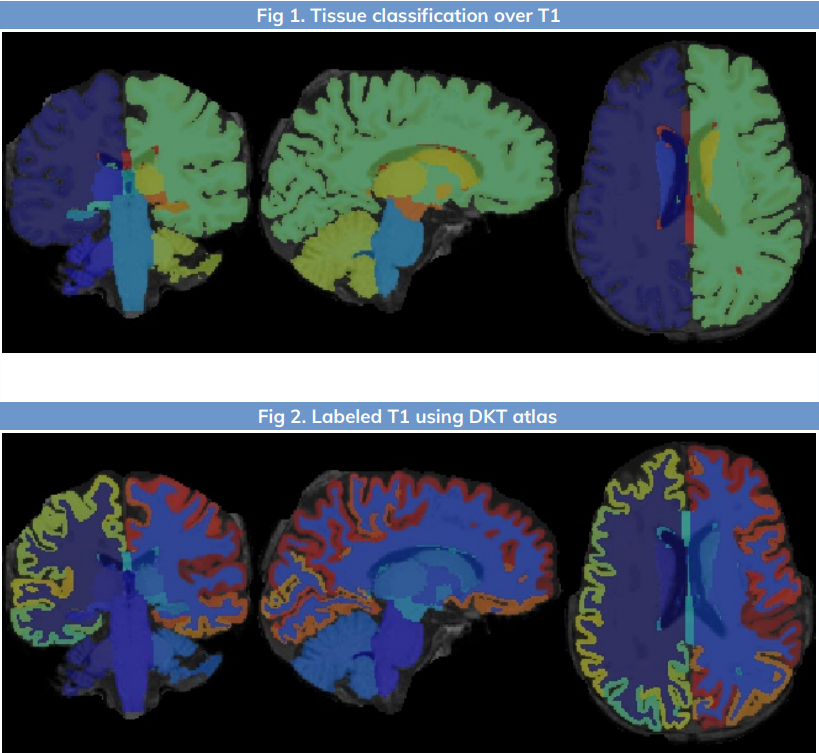
Fig 3 - Platform visualization of hippocampal subfields segmentation
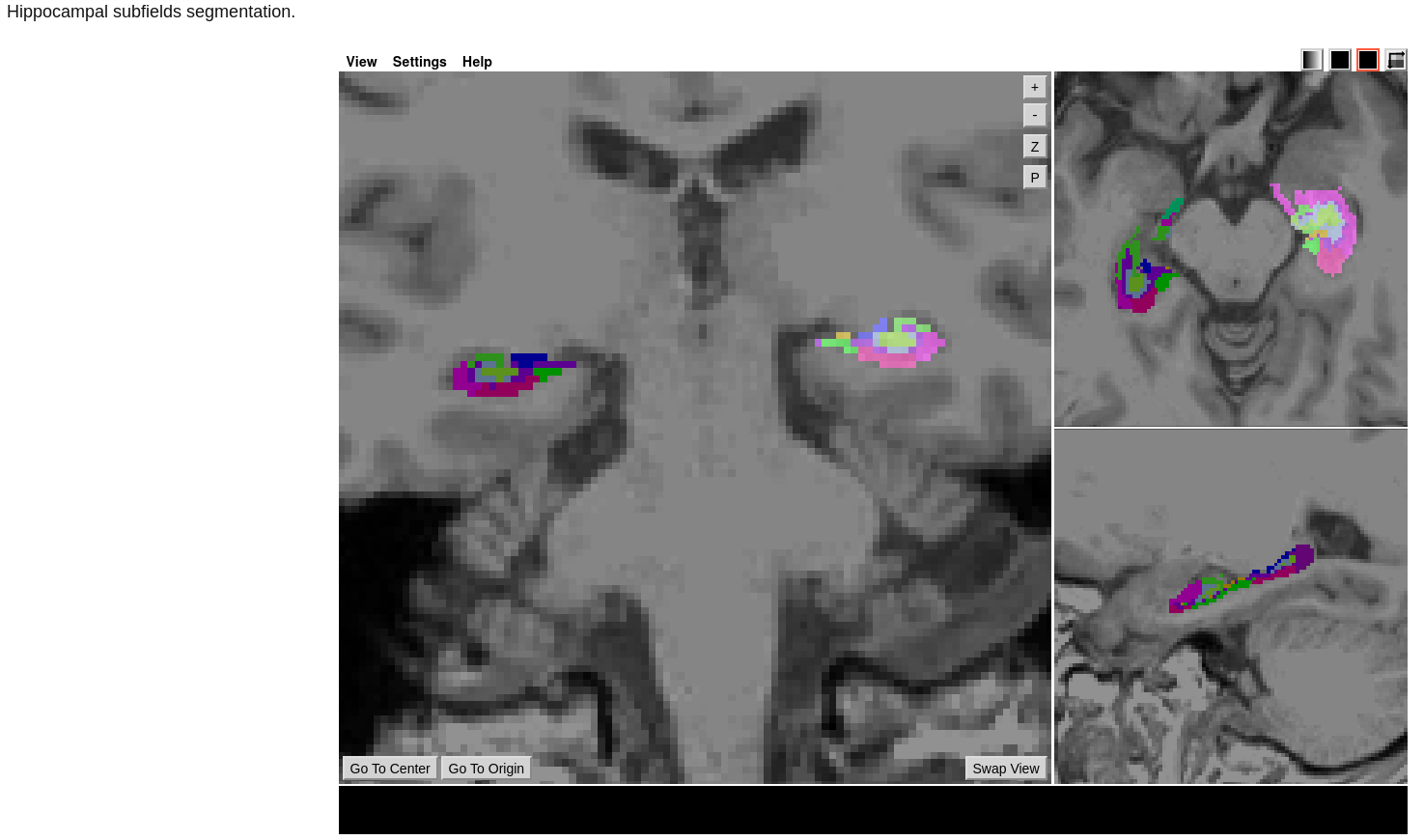
Fig 4 - Hippocampal subfields volumetric data
Required inputs:
- T1: anatomical 3D image
- Isotropic resolution recommended
- Must be labelled as 'T1' modality
Optional inputs:
- T2: anatomical 3D image
- Isotropic resolution recommended
- Must be labelled as 'T2' modality
- T2 FLAIR: anatomical 3D image
- Isotropic resolution recommended
- Must be labelled as 'T2' modality and have a ‘flair’ tag
- T2 (high-resolution): anatomical image covering hippocampi
Minimum input requirements:
- Image must contain full skull (not be brain extracted).For optimal result reliability, isotropic resolution is recommended
- Recommended resolution: 1 mm isotropic.
- Minimum reliable resolution: 2mm isotropic.
Outputs:
- Workflow main output:
- T1.nii.gz: the input T1 image used in the tool.
- Hippocampal_subfield_lh.nii.gz / hippocampal_subfield_rh.nii.gz: Hippocampal subfields segmentation files.
- Hippocampal_subfields_volumes.csv: Volumetric information about hippocampal region volumes.
- PostRECONALL step:
- report.pdf: report file with results summary.
- Aseg_stats CSV files: Freesurfer volumetry and thickness complete results.
- Volumetric.csv: CSV file with gray matter parcellated regions volume and cortical thickness information
- Images in RECONALL step:
- FREESURFER: Folder with full outputs of the FreeSurfer recon-all script [More information on FreeSurfer outputs] .
- T1.nii.gz: the input T1 image used in the tool.
- T1brain.nii.gz: preprocessed T1 image acpc-aligned.
- labeled.nii.gz: preprocessed and skull-stripped T1 image.
- tissueSegmentation.nii.gz: 5-tissue (csf, gm, wm, brainstem, cerebellum) tissue segmentation image.
The tool will write the total left and right hippocampal volume in the session’s metadata.
Typical execution time:
- 9 hours.
References:
- FreeSurfer: [Dale et al. 1999] , [Fischl et al. 1999] , [Fischl et al. 2000] , [Fischl et al. 2002] , [Fischl et al. 2004]
- Hippocampal segmentation step: [Iglesias, J.E et al 2015 ]
License Information:
- Users of this tool should be in possession of a FreeSurfer license. Obtaining a license is free and comes in the form of a license.txt file.
- More information on the FreeSurfer license can be found here .
- Follow this link to obtain a license key.
Create free account now!
