Brain Tumor Segmentation Workflow
This workflow segments the main structures of gliomas from MR images in a reproducible and fully-automated manner. The workflow expects a post-contrast T1 with Gadolinium agent (T1-Gad), a T2 FLAIR and optionally a native T1, and then produces class-confidence maps (softmaps) for each glioma sub-region, a segmentation map with the final prediction, and a PDF report providing a visual overview of the segmentation, tumoral volume quantification and a selection of radiomic features.
The segmentation map consists of the following value to class mapping:
- 0: non-tumoral tissue and background
- 1: edema
- 2: enhancing tumor
- 3: non-enhancing tumor and necrosis
The brain tumor segmentation tool included in the workflow is primarily made of a Convolutional Neural Network trained on a large proprietary database of pre- and post-operative MRI scans of patients with gliomas of varying shape, extension, and composition.
It includes an image pre-processing pipeline that resamples, co-aligns and skull-strips the input images to be prepared for inference with the aforementioned model. The model generates confidence-score maps for each tumoral sub-region and the final prediction map is obtained by assigning to each voxel the class with the highest confidence. No post-processing is applied after inference.
Afterward, the segmentation map is used to compute volumes and ratios of volumes of the different regions of interest. The segmentation map is also used, together with the post-contrast T1 image, to compute the following radiomic features:
- First Order statistics
- Shape features
- Gray Level Cooccurrence Matrix (GLCM)
- First Order statistics over Laplacian of Gaussian (LoG) with different sigma values (1mm, 3mm and 5mm).
All these volumetric and radiomic features are included in the PDF report.
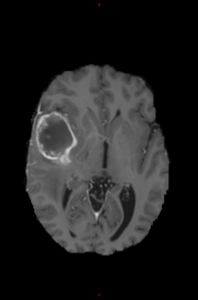
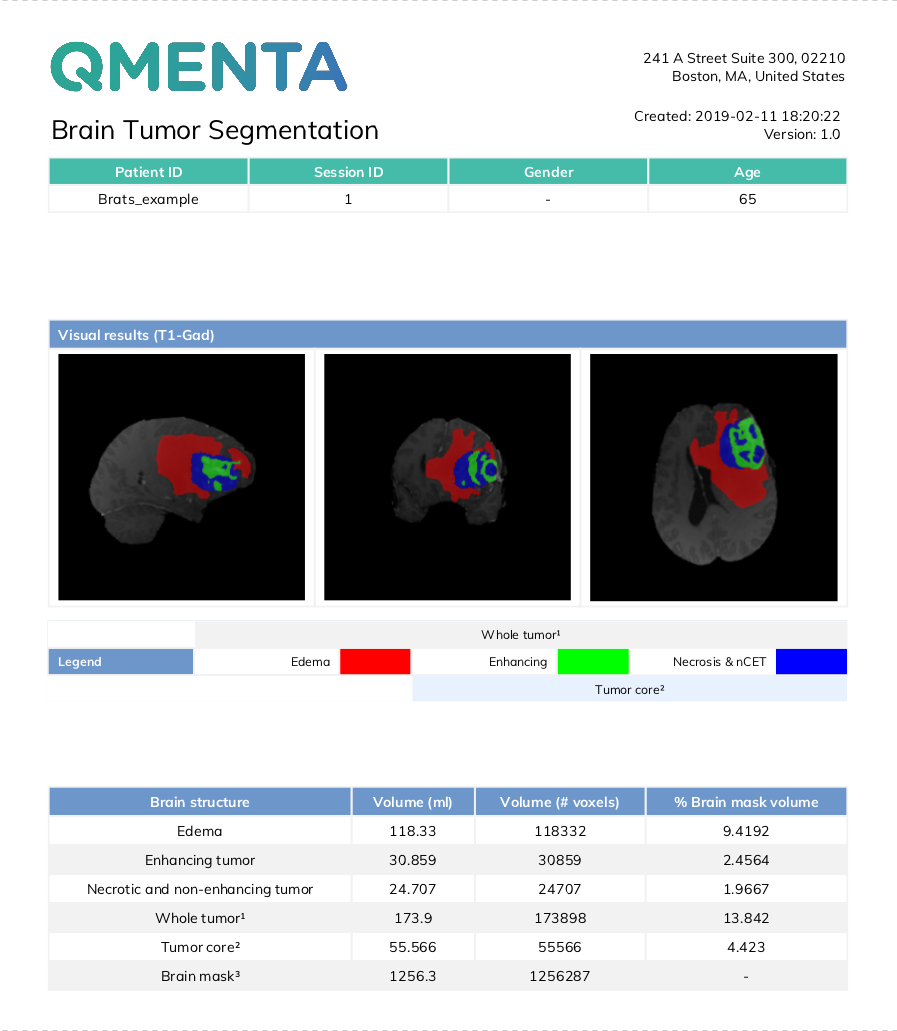
T1-Gad |
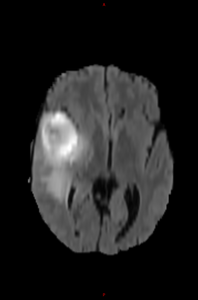
T2-FLAIR |
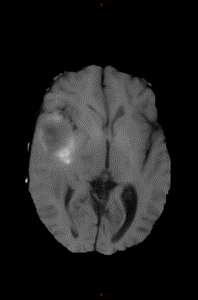 T1 (optional) |
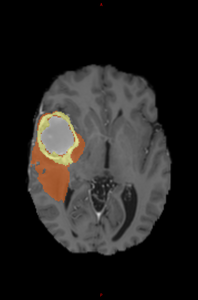
Segmentation map |
Required inputs
- T1 Gad: anatomical T1 post-contrast. Must be labelled as 'T1' modality and 'gad' tag.
- FLAIR: T2 FLAIR image. Must be labelled as 'T2' modality and 'flair' tag.
Optional inputs
- T1: T1 pre-contrast. Including a T1 pre-contrast image helps in the segmentation of necrotic tissue.
Must be labelled as 'T1' modality and have no tags.
Minimum input requirements
- Whole-brain FOVs are recommended for all input modalities.
- 3D images are highly recommended. The preferred resolution is 1mm isotropic in all cases.
- If resampling and co-registration pre-processing steps are skipped, the input images must have the same resolution and voxel dimensions.
Settings
-
Image pre-processing
-
Isotropic resampling to 1mm (checkbox). Controls whether isotropic resampling to 1mm is executed or not. As the model was trained on images with such resolution, it is compulsory to have the inputs passed to the model at such resolution. Therefore, this option should be unchecked only when the input image has already 1mm isotropic resolution.
-
Co-registration of image modalities (checkbox). Controls whether the co-registration of input images is run or not. This step should be skipped only when the input images are already co-aligned, in order to save some processing time.
-
Skull stripping (checkbox). Controls whether the skull-stripping step should be run or not. The segmentation network was trained exclusively with skull-stripped data, so the inclusion of input images with skull would introduce confounding information that might worsen the results provided by the tool.
-
Output container files
- MASKBRAIN brain_mask.nii.gz: Extracted brain mask, used for skull-stripping.
- SOFTMAP edema.nii.gz: Pseudo-probability map (softmap) of edema.
- SOFTMAP enhancing_tumor.nii.gz: Pseudo-probability map (softmap) of enhancing tumor.
- SOFTMAP necrosis.nii.gz: Pseudo-probability map (softmap) of necrotic tissue.
- LABELS prediction.nii.gz: Final segmentation map, with edema (1), enhancing tumor (2) and necrosis (3).
- report.pdf: PDF report.
- T1 GAD t1_gad.nii.gz: Pre-processed T1 Gadolinium contrast used to perform inference on the DL model.
- T2 FLAIR t2_flair.nii.gz: Pre-processed T2-FLAIR used to perform inference on the DL model.
- T1 t1.nii.gz (optional): Pre-processed T1 used to perform inference on the DL model.
- preprocessing/
- t1_gad_aligned.nii.gz: Pre-processed T1 Gadolinium contrast before skull-stripping. Used for visualization of results.
- t2_flair_aligned.nii.gz: Pre-processed T2-FLAIR contrast before skull-stripping. Used for visualization of results.
- t1_aligned.nii.gz (optional): Pre-processed T1 before skull-stripping. Used for visualization of results.
- radiomics/
- LOG LoG_sigma_1mm_t1_gad.nii.gz: Laplacian of Gaussian of T1-Gad image with sigma=1mm.
- LOG LoG_sigma_3mm_t1_gad.nii.gz: Laplacian of Gaussian of T1-Gad image with sigma=3mm.
- LOG LoG_sigma_5mm_t1_gad.nii.gz: Laplacian of Gaussian of T1-Gad image with sigma=5mm.
- report/
- online_summary_report.html: Online report, visible in the fourth tab of "Show results".
- online_radiomics_report.html: Online report, visible in the fourth tab of "Show results".
- tumor_segmentation_over_reference_axial.png: Axial view of multiplanar representation of the segmentation map.
- tumor_segmentation_over_reference_coronal.png: Coronal view of multiplanar representation of the segmentation map.
- tumor_segmentation_over_reference_sagittal.png: Sagittal view of multiplanar representation of the segmentation map.
- tables/
- VOLUMES tumor_volumes.csv: CSV with the volume quantification of the different tumoral regions of interest.
- RATIOS tumor_volumes_ratios.csv: CSV with the volume ratios of the different tumoral regions of interest.
- RADIOMICS radiomic_features.csv: CSV with the extracted radiomic features.
References
- ANTs registration: Avants et al. 2008
- ROBEX skull-stripping: Iglesias et al. 2011
- CNN architecture: Puch et al. 2019
- Radiomic features: van Griethuysen et al. 2017
Create free account now!
