Radiomics Workflow
This workflow takes a 3D medical image and a mask with one or more labels and extracts a series of radiomics features using the PyRadiomics library. It is especially thought for, but not limited to, oncology images characterization. The user can specify which radiomic classes want to compute and also optionally apply filters to the original image. If image filters are selected, radiomic features are also extracted from them. The computed radiomic features for each label in the mask are outputted in csv files, one per the original image and an additional one for each filter selected. The results are summed up into a structured report that can either be downloaded or viewed in the platform.
 |
 |
 |
The following radiomic classes can be selected:
- First Order Features
- Shape Features
- Gray Level Co-occurrence Matrix (GLCM)
- Gray Level Size Zone Matrix (GLSZM)
- Gray Level Run Length Matrix (GLRLM)
- Neighbouring Gray Tone Difference Matrix (NGTDM)
- Gray Level Dependence Matrix (GLDM)
The following image filters can be selected:
- Wavelet
- Laplacian of Gaussian (Needs extra parameters to be set)
- Sigma
- Filter width
- Logarithm
- Exponential
The workflow includes
- DICOM files to NIfTI conversion and reorients to standard. The workflow file filter sends the dicom or nifti files required for the workflow to run and the box converts the dicom to niftis.
- Computation of radiomic features for each label in the input mask. The radiomic classes can be chosen.
- If image filters are selected, the original image is filtered and radiomic features are computed over the result.
- All the nifti files and csv files are collected and a report with the most relevant results is created. The report can be downloaded as a pdf and also be displayed in the platform.
Required inputs:
-
Medical image:
- Can be either a NIfTI or DICOM file (DICOM files will be converted before radiomics extraction).
- Accepted modalities: T1, T2, CT and SCALAR.
-
Labels mask:
- Must be in the same space as the medical image.
- Can contain one or more labels.
- Must be tagged as labels, mask or both.
Minimum input requirements:
-
For optimal result reliability, an isotropic resolution is highly recommended.
- Recommended resolution: 1mm isotropic.
Output files:
-
DCM2NII:
- Oncology medical image in NIfTI format.
- Labels mask in NIfTI format
- Report for quality control and visualization
-
Radiomics:
- Original input image in NIfTI format.
- Filtered images in NIfTI format for each image filter selected.
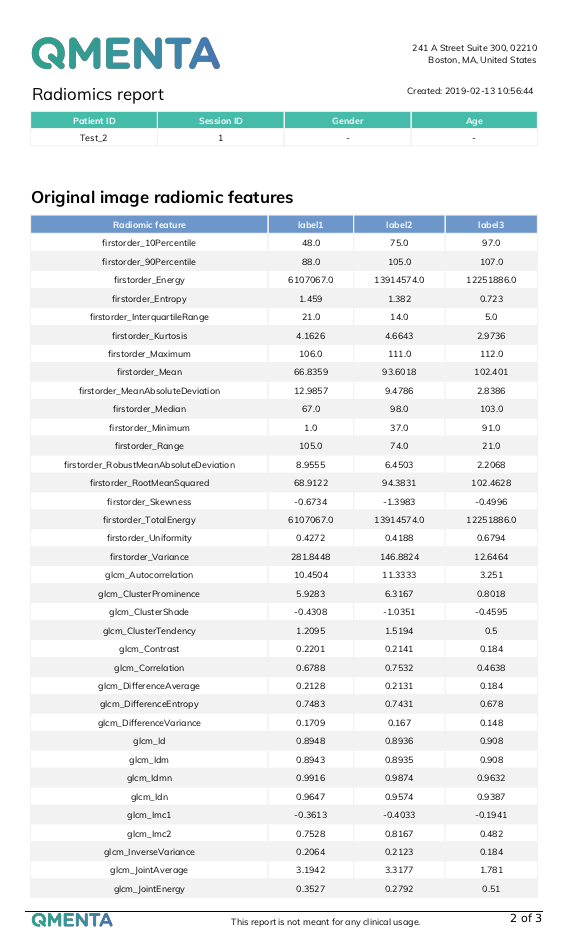
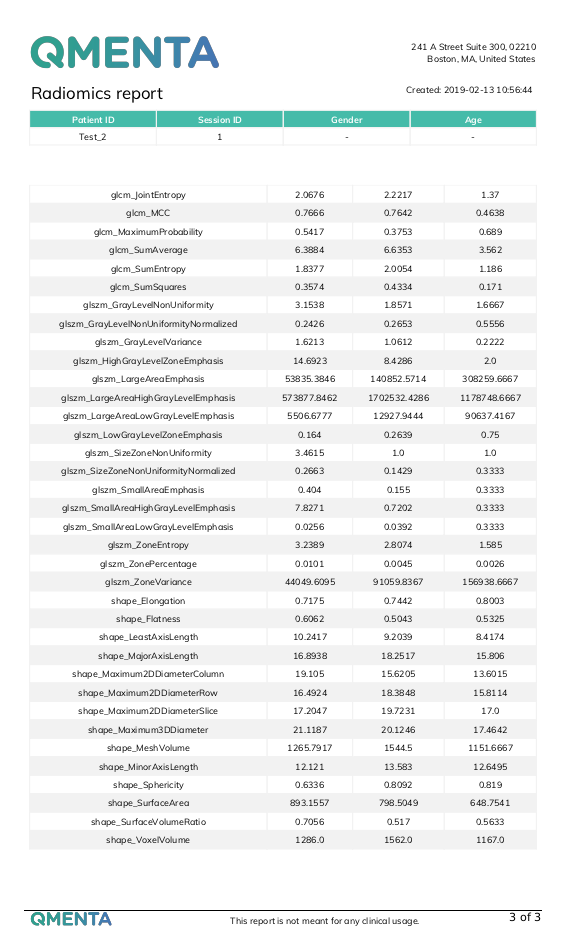
- A csv file with radiomic features for each label in the original image.
- An extra csv file for the radiomics of each filtered image.
- Labels mask in NIfTI format.
- Original input image in NIfTI format.
-
Radiomics report:
- All output files from the Radiomics tool divided in folders.
- PDF report collecting all the relevant results.
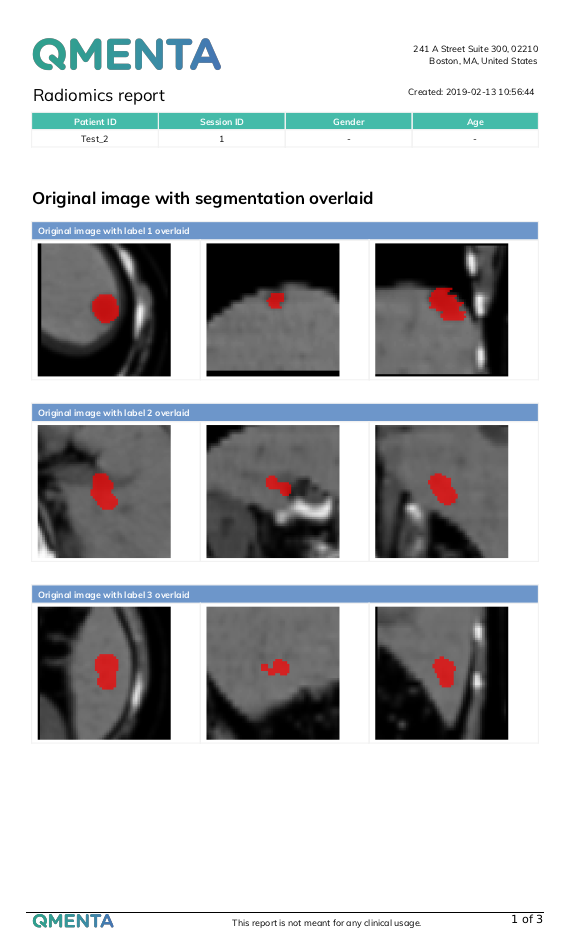
- PNG files showing the labels overlaid to the original and filtered (if applicable) images from three different orientations.
- An html report that can be displayed by selecting the Show results option in a finished workflow.
References
Create free account now!
