Intensity-based segmentation of main tumor mass from a T2 or T2-FLAIR image. input image will be resampled to 1mm isotropic.
Atlas-based information is used to analyze the tumor damage by region. CSF prior information is used to exclude CSF from non-FLAIR input images.
Quantification outputs with percentage of region overlap are written to a report PDF and a CSV datasheet.
Tumor Segmentation Results 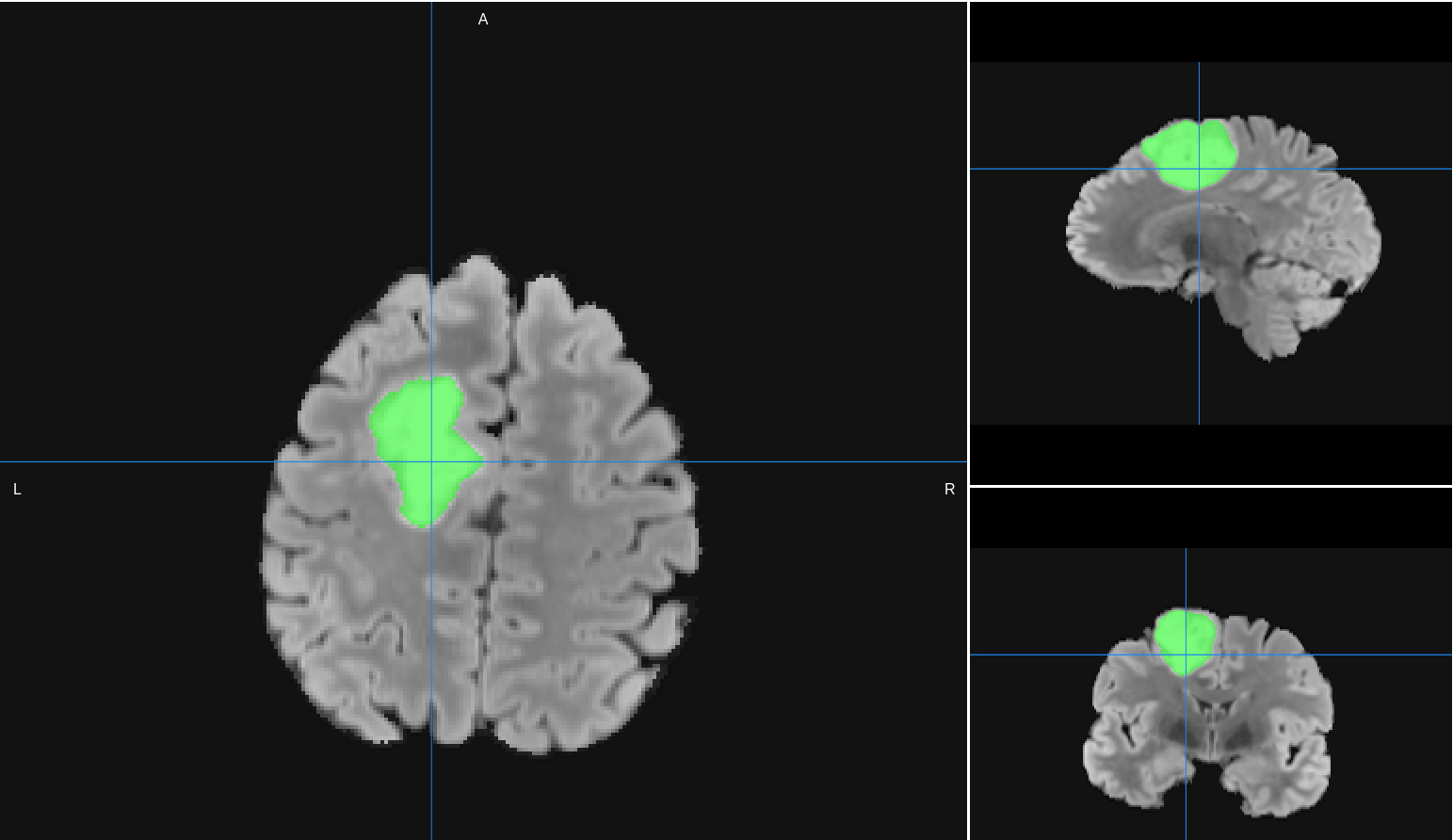
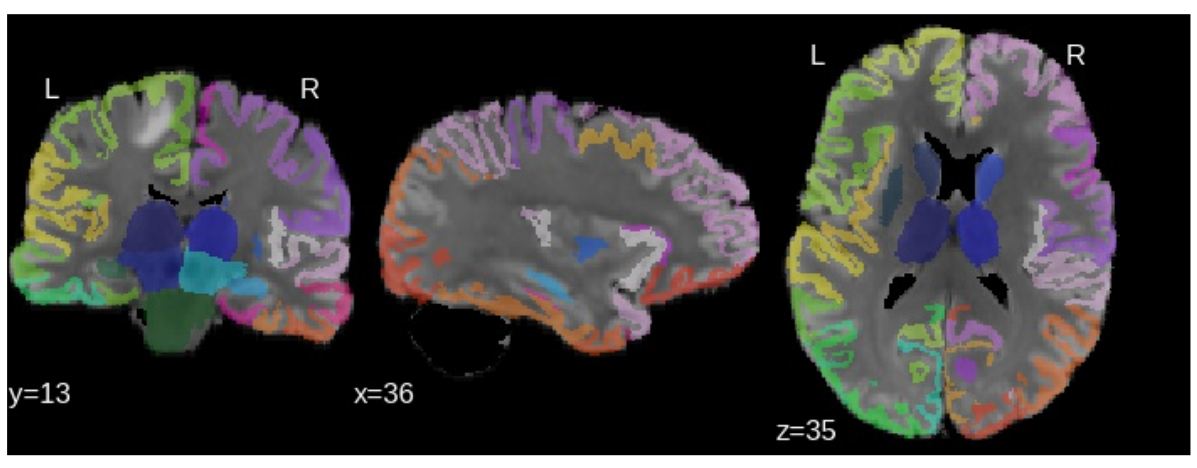
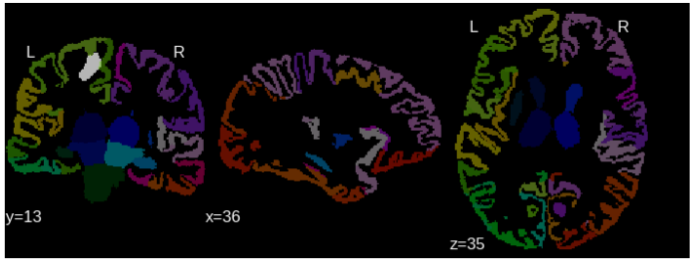
Affected regions 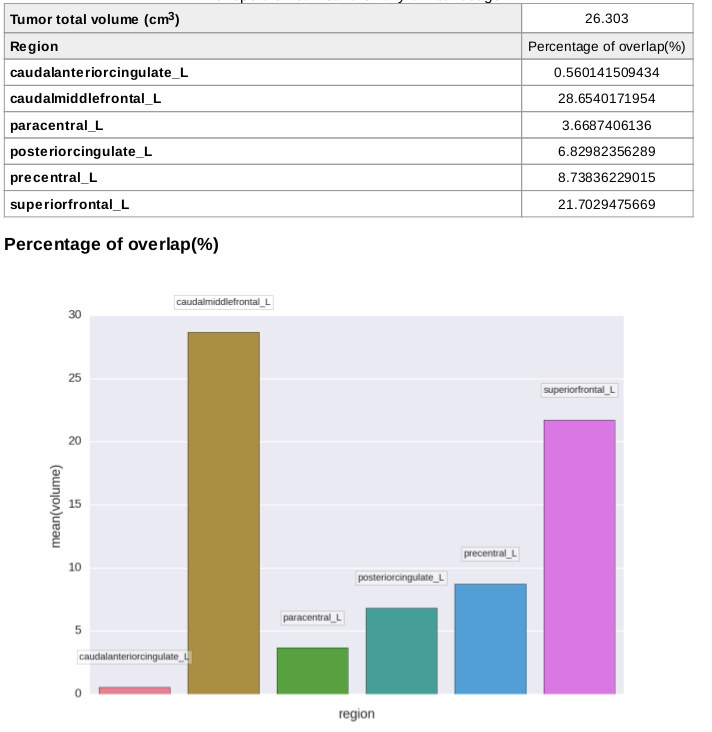
- Inputs:
- A T2 image, image file must be labelled as "T2" modality.
- Minimum input requirements:
- Image must contain full skull (not be brain extracted).
- For optimal result reliability, isotropic resolution is recommended
- Recommended resolution: 1 mm isotropic.
- Minimum reliable resolution: 2mm isotropic.
- Parameters:
- Preferred DICOM to Nifti conversion tool.
Tool used to convert from DICOM to NIfTI.- Mrtrix (Default)
- DCM2nii
- diffunpack
- mcverter
- Skull Strip Parameters:
Skull stripping is customizable in the BrainSuite (please refer here for more details) and AFNI's 3dSkullStrip case using different options available in its software (please read carefully the parameter definition before using it).
- Tool for skull-stripping:
Tool used to skull strip the T2w image converted to NIfTI- AFNI (Default)
- BET
- Brainsuite
- Parameters
String with extra parameters for the skull-stripping tool- Type: String
- Default: ''
- Tool for skull-stripping:
- Atlas selection:
Which atlas to use,- Type: single choice
- AAL (default)
- Registration tool:
Tool used to register the template to the subject brain- ANTs (Default)
- FSL
- Tumor segmentation parameters:
- Threshold for masking tumor:
Percentile of the image where to threshold, we keep the values above that percentile- Type: decimal
- Default: 99
- Size of the correction:
Size of the erosion performed to clean the data before removing small objects and then dilate by the same- Type: decimal
- Default: 3
- Threshold for masking tumor:
- Preferred DICOM to Nifti conversion tool.
- Outputs:
- Images:
- T2_corr_resampled.nii.gz: Corrected T2 image (skull strip and resampling)
- T2_corr_resampled_no_csf.nii.gz: CSF cleaned T2
- T2_labeled.nii.gz: Labels registered to the T2 image.
- T2_labeled_with_tumor.nii.gz: Highlighted regions of the atlas overlapping with tumor.
- T2_tumor.nii.gz: Segmented tumor binary image
- Registration step:
- template0GenericAffine.mat: Transformation matrix used for atlas registration
- templateWarped.nii.gz: Intermediate registration result using brain template
- Datasheets:
- volumetric_tumor.csv: CSV with the tumor analysis information
- Report:
- report.pdf
- Images:
- Typical execution time:
- 1h25
- 1h25
-
- ANTs script: Neuroimage
- ANTs registration: Medical image analysis
Create free account now!

